Mimicking GEFs: a common theme for bacterial pathogens
Summary
Small molecular weight GTPases are master regulators of eukaryotic signalling, making them prime targets for bacterial virulence factors. Here, we review the recent advances made in understanding how bacterial type III secreted effector proteins directly activate GTPase signalling cascades. Specifically we focus on the SopE/WxxxE family of effectors that functionally mimic guanine nucleotide exchange factors (GEFs): the endogenous activators of Rho-family GTPases. Recent structural and biochemical studies have provided keen insight into both the signalling potency and substrate specificity of bacterial GEFs. Additionally, these bacterial GEFs display fascinating cell biological properties that provide insight into both host cell physiology and infectious disease strategies.
Introduction
Many Gram-negative bacterial pathogens use a type III secretion system (TTSS) to deliver bacterial effector proteins directly into host cells. These effector proteins are potent regulators of the intracellular signalling environment, thus providing bacteria a direct mechanism to control host cell morphology, dynamics, and metabolism (Cornelis, 2006; Galan, 2009). It is now clear that small molecular-weight GTPases are critical targets of bacterial effector proteins. These enzymes are responsible for controlling signal transduction pathways, membrane trafficking, cytoskeletal dynamics, nuclear import, and a wide range of physiological processes. In order to carry out these functions, G proteins are rapidly recruited from cytoplasm to membranes in response to extracellular stimuli. Once localized, they are activated by the classic switching mechanism involving GTPase conversion from GDP-bound inactive to the GTP-bound active state. Facilitating the conversion of these states are two groups of proteins: guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs). GEFs activate G-proteins by catalysing the exchange of GDP for GTP. Conversely, GAPs function by binding and accelerating the slow intrinsic hydrolysis rate of the G-proteins, thus inactivating the protein (Bos et al., 2007). A large body of work now shows that pathogenic bacteria have evolved intricate mechanisms to inhibit G-protein signalling through a range of strategies including proteolysis and post-translational modifications (Sekine et al., 1989; Flatau et al., 1997; Shao et al., 2002; Yarbrough et al., 2009). In this article we explore the alternative pathway of GTPase regulation, namely activation by guanine nucleotide exchange. Specifically, we focus on the SopE/WxxxE family of bacterial type III effectors that mimic the activity of eukaryotic GEF proteins.
Discovering bacterial GEF mimicry
The Salmonella type III effector protein SopE was the first reported bacterial GEF. Hardt et al. discovered that SopE directly activates Cdc42 and Rac GTPases to induce membrane ruffling at the site of Salmonella invasion (Hardt et al., 1998). The discovery lead to the identification of several additional bacterial GEFs that displayed genetic similarity to SopE, including SopE2 (Salmonella spp.), BopE (Burkholderia pseudomallei) and CopE (Chromobacterium violaceum). Until recently, this family of type III effectors was the only Rho-specific GEFs to be identified in bacterial species.
In 2006, Alto et al. identified a bacterial effector family that activated GTPase signalling cascades within host cells (Alto et al., 2006). While this family shares very low sequence homology (< 15%) they all contain an invariant Trp-X-X-X-Glu signature motif (Alto et al., 2006). The original description of the so-called ‘WxxxE’ family (pronounced whi-xee) suggested that these effector proteins directly mimic small GTPases (Alto et al., 2006). However,it was clear from subsequent structural studies that the WxxxE effectors adopt a GEF-like fold similar to SopE (Ohlson et al., 2008). This finding was quite surprising since sequence-based alignment of the WxxxE and SopE effector does not yield any significant homology. Huang et al. reported the first GEF activity for three WxxxE family members: Map, IpgB1 and IpgB2 (Huang et al., 2009). Additional reports have confirmed that the WxxxE proteins function as GEFs and not as GTPase mimics (Arbeloa et al., 2010; Klink et al., 2010). Importantly, the structural, pathogenic and cellular regulation of the SopE/WxxxE family of bacterial GEFs has greatly increased our understanding of bacterial GEF mimicry at the molecular level.
Catalytic mechanism of GEF mimics
Structural studies investigating the SopE/WxxxE family have been instrumental in understanding the underlying GTPase activation mechanism utilized by bacterial GEFs. Currently, there are solved structures for the GEFs SopE, SopE2, BopE, Map, SifA and IpgB2 (Buchwald et al., 2002; Upadhyay et al., 2004; Williams et al., 2004; Ohlson et al., 2008; Huang et al., 2009; Klink et al., 2010). Buchwald et al. solved the structure of SopE in complex with Cdc42, providing the first structural insight into bacterial GEF mimics (Buchwald et al., 2002). Surprisingly, SopE does not resemble the Dbl-homology (DH) domain or the Dock Homology Region 2 (DHR-2) domain, the two major classes of eukaryotic Rho GEFs (Fig. 1A). Rather, SopE adopts a V-shaped fold consisting of two bundles of alpha helices and an extended catalytic loop that connects the two helical bundles (Fig. 1A) (Buchwald et al., 2002). Closer inspection of the interactions between SopE and Cdc42 does reveal general similarities of guanine nucleotide exchange between bacterial and eukaryotic GEFs. It appears that all GEFs interact extensively with the GTPase switch 1 and switch 2 loops (Buchwald et al., 2002). The GTPase switch loops define the major structural differences between an inactive (GDP-bound) and active (GTP-bound) state (Vetter and Wittinghofer, 2001). More specifically, both SopE and the Dbl family of GEFs utilize an acidic residue and an amide side chain to interact with residues on switch 1 and switch 2 respectively (Buchwald et al., 2002). Recent investigations on the WxxxE family of GEFs illustrates that they too adopt a V-shaped structure similar to SopE (Fig. 1A) (Ohlson et al., 2008; Huang et al., 2009). Additionally, the WxxxE proteins utilizes the conserved acidic and amide residues to interact with the GTPase switch loops similar to SopE and Dbl GEFs (Huang et al., 2009). Lastly, both SopE and WxxxE bacterial GEFs induce nearly identical conformational changes around the GTPase nucleotide binding site as the Dbl family of eukaryotic GEFs (Buchwald et al., 2002; Huang et al., 2009). These structural studies have confirmed that both the WxxxE and SopE GEF mimics have converged upon a similar guanine nucleotide exchange mechanism.
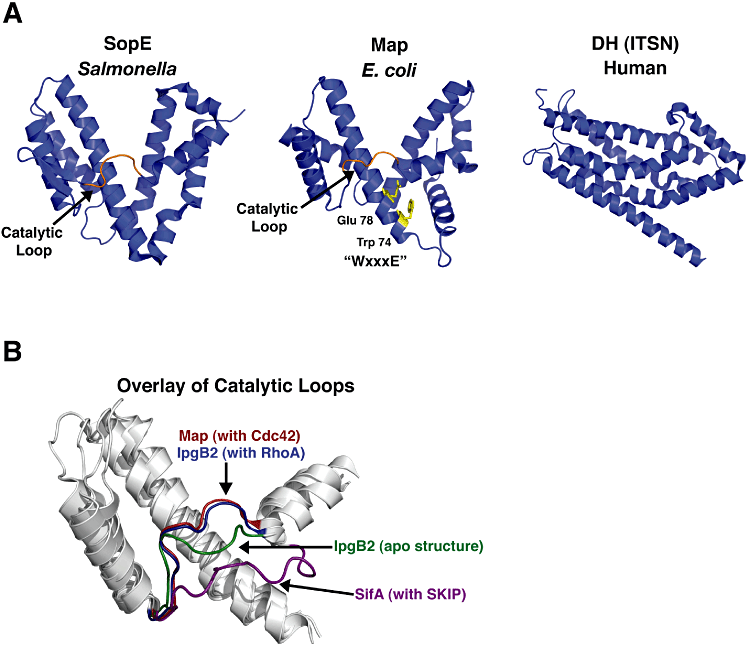
Structural comparison of bacterial GEF mimics. A. Structure of Map from E. coli (PDB: 3GCG), SopE from Salmonella (PDB: 1ZGS), and the human GEF ITSN (PDB: 1KIL). Highlighted in orange is the catalytic loop of Map and SopE. The side-chain of the Tryptophan and Glutamic acid of Map's WxxxE motif are shown as yellow sticks. B. Comparison of the catalytic loops of the WxxxE GEFs. Structures of indicated GEF constructs were overlayed to illustrate the different position of the loops. Map in complex with Cdc42 (red; PDB: 3GCG) and IpgB2 in complex with RhoA (blue; PDB: 3LXR) lie higher on the V structure compared with the loops of uncomplexed IpgB2 (green; PDB: 3LYQ) and SifA (purple; PDB: 3CXB).
The catalytic loop of these bacterial GEF mimics is important for making contacts with the GTPase switch 1 and switch 2 regions (Buchwald et al., 2002; Huang et al., 2009). Recent studies have demonstrated that the catalytic loop is flexible and that proper orientation is important for GTPase recognition and activation (Klink et al., 2010). For example, in the structures of Map and IpgB2 in complex with Cdc42 and RhoA, respectively, the catalytic loop sits high upon the V fold (Fig. 1B). Further examination of IpgB2 shows that the catalytic loop lies much lower on the V structure in the apo-structure compared with when IpgB2 is in complex with RhoA (Fig. 1B). Similarly, in the structure of SifA in which there is no GTPase present, the catalytic loop of SifA lies even lower on the structure (Fig. 1B). It now appears that reorientation of the catalytic loop may be a mechanism of regulating GEF activity (Klink et al., 2010). Understanding the functional consequences of undergoing such conformational changes remains an outstanding question in the field.
Bacterial GEF mimics also provide keen insight into how eukaryotic GEFs function. For example, Klink et al. provide new insights into the GTPase activation mechanism of bacterial GEFs, which has implications for endogenous GEFs as well (Klink et al., 2010). The structure of IpgB2 in complex with RhoA was solved under three different magnesium concentration conditions. With mild treatment of EDTA excess Mg2+ ions were depleted from the complex and the solved structure showed a novel Mg2+ binding site for RhoA (Klink et al., 2010). Typically, Mg2+ is found near the β-phosphate of the GDP but at low Mg2+ concentrations, the Mg2+ relocates to the α-phosphate position of the nucleotide (Klink et al., 2010). This new secondary Mg2+ site provides new insight into GTPase activation through the stepwise transition of GDP for GTP.
Selecting appropriate GTPase substrates
Structural studies have also been fundamental to understanding the mechanisms of GTPase isoform selection by bacterial GEFs. In their original description, Alto et al. showed that WxxxE proteins activate specific Rho signalling cascades, but the mechanism of discrimination was unknown (Alto et al., 2006). For example, the protein Map is a highly specific GEF for Cdc42 and does not activate Rac1 or RhoA isoforms (Huang et al., 2009). Examining the structure of Map in complex with Cdc42 has revealed important interactions between Map and the β2–3 inter-switch region of Cdc42 (Huang et al., 2009). The β2–3 region is significant since it is the most variable region in the Rho family GTPases and is utilized by the eukaryotic Dbl family of GEFs to distinguish between different GTPase isoforms (Snyder et al., 2002). Indeed, the specificity of Map can be changed from Cdc42 to Rac by simply inter-converting the β2–3 region of these GTPases (Huang et al., 2009). While the β2–3 region is the discriminatory element on the GTPases, the bacterial GEFs also have variable features that recognize this element. A structural alignment of the WxxxE family revealed that the amino acids that interact with the β2–3 region are highly variable in the WxxxE proteins, yet are conserved among GEFs that select the same GTPase isoforms (Huang et al., 2009; Klink et al., 2010). Therefore, the GEF mimicry goes beyond just the GTPase activation mechanism, but the specificity mechanism as well. The SopE subfamily of GEFs does not engage the β2–3 region like the WxxxE subfamily, which raises the question of how these GEFs discriminate between various GTPase isoforms (Buchwald et al., 2002; Huang et al., 2009).
The regulation and function of bacterial GEFs in pathogenesis
The pathogenic role of the SopE/WxxxE bacterial GEFs has been extensively studied, but the mechanisms to regulate eukaryotic signalling pathways are still emerging. It now appears that similar to eukaryotic GEFs, the SopE/WxxxE bacterial GEF mimics utilize complicated lipid and protein interactions to regulate their activity. Also, some pathogens encode multiple bacterial GEFs that differ in substrate specificity, raising the question of how these proteins work in context of numerous bacterial infection mechanisms. A summary of each GEF's biochemical activity is listed in Table 1.
| Species | Protein | Family | Substrate(s) | Bindinga | GEF activitya | Structure (PDB) | References |
|---|---|---|---|---|---|---|---|
| Salmonella | SopE | SopE | Rac1 & Cdc42 | Yes | Yes | 1GZS | Hardt et al. (1998) |
| SopE2 | SopE | Rac1 & Cdc42 | Yes | Yes | 1R9K | Friebel et al. (2001) | |
| SifA | WxxxE | RhoA | Yes | None Detected | 3HW2 3CXB | Ohlson et al. (2008) | |
| SifB | WxxxE | Unknown | Unknown | Unknown | Alto et al. (2006) | ||
| Burkholderia | BopE | SopE | Rac1 & Cdc42 | Yes | Yes | 2JOK 2JOL | Upadhyay et al. (2004) |
| Shigella | IpgB1 | WxxxE | Rac1 & Cdc42 | Yes | Yes | Huang et al. (2009) | |
| IpgB2 | WxxxE | RhoA | Yes | Yes | 3LW8 3LWN 3LXR 3LYQ | Huang et al. (2009; Klink et al. (2010) | |
| A/E lesion pathogens | Map | WxxxE | Cdc42 | Yes | Yes | 3GCG | Huang et al. (2009) |
| EspM | WxxxE | RhoA | Yes | Yes | Arbeloa et al. (2010) | ||
| EspT | WxxxE | Rac1 & Cdc42 | Unknown | Unknown | Bulgin et al. (2009b) | ||
| Chromobacterium violaceum | CopE | SopE | Rac1 & Cdc42 | Unknown | Unknown | Miki et al. (2011) |
- a. Binding and GEF activity refer to whether or not biochemical studies have confirmed binding and nucleotide exchange of GTPase substrates.
Salmonella's GEFs
Salmonella spp. encodes four bacterial GEFs, two SopE-type and two WxxxE-type. SopE and its homologue SopE2 are secreted by the SPI-1 TTSS and are required for the entry of Salmonella into non-phagocytic cells (Fig. 2A) (Hardt et al., 1998; Bakshi et al., 2000; Stender et al., 2000). These GEFs induce membrane ruffles required for Salmonella invasion through the activation of Rac and Cdc42 (Hardt et al., 1998; Bakshi et al., 2000; Stender et al., 2000). While SopE and SopE2 have slightly different substrate preferences with SopE activating both Cdc42 and Rac1- and SopE2-activating Cdc42 (Friebel et al., 2001), Rac1 activation is the primary driving factor for Salmonella entry into host cells (Patel and Galan, 2006). However, given that Cdc42 activates Rac1 through GTPase cross-talk, either GEF is sufficient to induce host cell invasion.
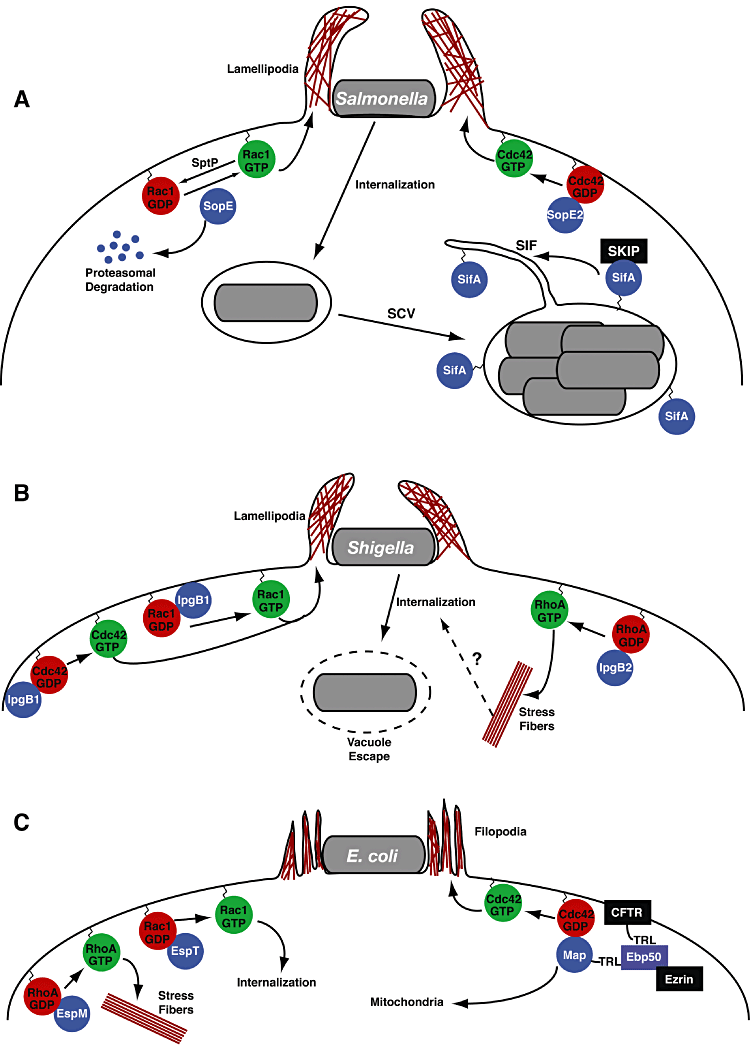
Bacterial GEFs in pathogenesis. Illustration of the function of bacterial GEFs (blue circles) in Salmonella (A), Shigella (B) and E. coli (C) pathogenesis. The GEFs, SopE, SopE2, IpgB1, IpgB2 and Map are expressed early during infection to generate specific actin structures. SifA is expressed later during Salmonella infection and is responsible for maintaining the Salmonella-containing vacuole (SCV). Pathways are further described in the text.
Paradoxically, Salmonella utilizes SopE to induce the host inflammatory response. SopE activates caspase-1, which processes the pro-inflammatory cytokines IL-1 and IL-18 (Muller et al., 2009). Surprisingly, transfection of SopE alone but not a mutant deficient in GEF activity is sufficient to activate caspase-1 (Muller et al., 2009). This inflammatory stimulation provides an unusual mechanism for Salmonella to outcompete the natural microbiota of the intestine (Stecher et al., 2007). Therefore, SopE not only functions in the invasion of host cells but also aids in the colonization of host tissues. It is tempting to speculate that the other pathogens that harbour SopE/WxxxE GEF mimics may also induce inflammation in addition to their currently known functions.
The cellular invasion mechanism utilized by Salmonella also provides a good example for the complex regulation of bacterial GEFs inside eukaryotic hosts. After SopE induced ruffle formation and invasion, a second type III effector SptP remodels the actin cytoskeleton by inactivating GTPases. SptP functions as a GAP that directly antagonizes the activation of Rac1 and Cdc42 by SopE (Fig. 2A) (Fu and Galan, 1999). In order to coordinate the timing of SopE and SptP activity, Salmonella utilizes the host proteasome degradation pathway (Fig. 2A). The amino terminus of SopE contains an ubiquitination motif that facilitates the rapid degradation of the SopE protein compared with SptP (Kubori and Galan, 2003). By generating chimeric proteins that alter SopE's and SptP's half-life, Kubori and Galan (2003) were able to demonstrate the importance of temporally regulating SopE and SptP activities during Salmonella infection. Adding to the complexity of the regulation of SopE and SptP is a recent report that the translocation rates of these two proteins are significantly different (Van Engelenburg and Palmer, 2008). Whether other bacteria fine-tune the temporal activity of their GEF mimics through the secretion of antagonizing effectors is an interesting but currently unexplored area of research.
As mentioned above, Salmonella also encodes two additional WxxxE type GEF mimics: SifA and SifB. Both SifA and SifB are secreted by the SPI-2 TTSS. While very little is known about SifB, SifA is required for full virulence in macrophages and in mice (Beuzon et al., 2000). SifA is secreted after Salmonella internalization and is necessary for the maintenance of the Salmonella-containing vacuole (SCV) and generation of Salmonella induced filaments (Sifs; Fig. 2A) (Beuzon et al., 2000). Without SifA, Salmonella are no longer able to control the endocytic trafficking to the SCV and bacteria escape out of the SCV (Beuzon et al., 2000; Brumell et al., 2002; Ruiz-Albert et al., 2002). SifA is anchored to the SCV via a C-terminal CaaX box that becomes lipidated (Boucrot et al., 2003; Reinicke et al., 2005). Without this modification on SifA, Salmonella are unable to maintain the SCV (Boucrot et al., 2003).
SifA is a two-domain protein with the N-terminus responsible for interacting with the host protein SKIP and the C-terminus adopts the SopE/WxxxE GEF (Ohlson et al., 2008). The interactions between SifA and SKIP are essential for maintenance of the SCV (Boucrot et al., 2005). SifA's interaction with SKIP is believed to link kinesin-1 activity with the events occurring on the SCV (Boucrot et al., 2005; Dumont et al., 2010). Intriguingly, the small GTPase Rab9 interacts with SKIP, and this interaction is disrupted by SifA (Jackson et al., 2008). Because Rab9 is involved in late endosomal trafficking, SifA's disruption of the Rab9::SKIP complex may function to antagonize Rab9's native functions (Jackson et al., 2008). Currently, there is not a known function of Rab9 in altering the SCV and the physiological implications of this finding still need to be explored.
Other genetic studies indicate that SifA requires another Salmonella effector, SseJ, to regulate membrane dynamics (Ruiz-Albert et al., 2002). SseJ is a member of the GDSL family of lipases/esterases and like other members of this family has a broad range of substrates (Ohlson et al., 2005). Deletion of both SifA and SseJ from the Salmonella genome allows bacteria to remain in the SCV (Ruiz-Albert et al., 2002). Additionally, SseJ and SifA appear to interact with each other in vivo (Ruiz-Albert et al., 2002; Ohlson et al., 2008). Because SseJ becomes more active in the presence of GTP-bound RhoA, it has been hypothesized that SifA functions as a RhoA GEF (Ohlson et al., 2008; Christen et al., 2009). Ohlson et al. have reported that SifA can interact with RhoA in the presence of HeLa cell lysate (Ohlson et al., 2008). However, the ability of SifA to catalyse nucleotide exchange on RhoA has not been demonstrated and structural modelling predicts highly unfavourable interactions between SifA and RhoA at the specificity determining region of RhoA (Klink et al., 2010). Because SifA is responsible for altering endomembrane trafficking, it is tempting to speculate that it may activate a small GTPase involved in the general secretory pathway such as Arf or Rab family GTPases (Beuzon et al., 2000; Ohlson et al., 2008). Future biochemical studies elucidating SifA's GTPase target will lend great insight into how Salmonella maintain the SCV.
Shigella's GEFs
Shigella spp. encode for two bacterial GEF mimics of the WxxxE family: IpgB1 and IpgB2 (Fig. 2B) (Alto et al., 2006; Huang et al., 2009). IpgB1 was initially identified as a type III effector required for efficient invasion of host cells (Ohya et al., 2005). Handa and colleagues presented evidence that IpgB1 recruits the ELMO/DOCK 180 complex to the membrane to activate Rac1 (Handa et al., 2007). Given the recent biochemical data demonstrating IpgB1's GEF activity, this finding needs to be confirmed, and could demonstrate a redundant mechanism to stimulate Rac1 activity. (Ohya et al., 2005; Huang et al., 2009). The N-terminus of IpgB1 associates with the plasma membrane through an unknown mechanism that may be important in regulating GEF catalysis (Handa et al., 2007).
Shigella's other bacterial GEF mimic, IpgB2, is less well understood. IpgB2 induces stress fibres in cells through activation of RhoA (Fig. 2B) (Huang et al., 2009). Shigella strains with the IpgB2 gene deleted are not deficient in cell invasion (Hachani et al., 2008). However, infection of polarized Caco-2 cells with single or double IpgB1/IpgB2 knockout strains produced some intriguing results. First, while IpgB1 deletion strains were defective for invasion into HeLa cells, the same strains had no defect in polarized cells (Hachani et al., 2008). A double knockout of IpgB1 and IpgB2 revealed a significant decrease in invasion of polarized epithelial cells and attenuation in a murine intranasal model (Hachani et al., 2008). These results, if confirmed, suggest that Shigella utilizes a more sophisticated means of invasion than previously appreciated. However, more extensive studies investigating the relationship between IpgB1, IpgB2 and host cell invasion are needed.
Attaching and effacing (A/E) lesion pathogen's GEFs
A/E pathogens include the closely related bacteria enteropathogenic E. coli (EPEC), enterohaemorrhagic E. coli (EHEC) and Citrobacter rodentium. Map, a WxxxE family GEF, is found in all of these pathogens. Map is a Cdc42-specific GEF that induces actin-filopodia protrusions around the infecting bacterium (Fig. 2C) (Kenny et al., 2002; Huang et al., 2009). Map was originally identified as a mitochondrial associated protein that interferes with the maintenance of the organelle's membrane potential (Kenny and Jepson, 2000). However, it is unclear whether the mitochondrial import signal is related to Map's activation of Cdc42. Map also harbours a PDZ-ligand that is important for its cellular regulation (Alto et al., 2006; Simpson et al., 2006). PDZ-ligands are short sequences at the C-termini of proteins that facilitate protein-protein interactions with proteins containing PDZ domains (Harris and Lim, 2001). Map's PDZ ligand is identical to that of the cystic fibrosis transmembrane regulator (CFTR) and like the CFTR, Map binds to the PDZ domains of Ezrin Binding Protein 50 (Ebp50) (Alto et al., 2006). The generation of filopodia protrusions by Map is dependent upon its ability to interact with Ebp50 (Fig. 2C) (Alto et al., 2006; Simpson et al., 2006). Why Map requires Ebp50 for signalling is still unclear. Currently, two hypotheses have been presented. First, Ebp50 might serve as a localization motif especially since it is known to associate with transmembrane proteins (Alto et al., 2006). Alternatively, Berger et al. suggest that Ebp50 is required for the stabilization of filopodia through the activation of RhoA (Berger et al., 2009). Future experiments directly testing these hypotheses are necessary.
In addition to Map, A/E lesion pathogens contain two other bacterial GEF mimics: EspM and EspT (both are WxxxE proteins). In a recent survey of clinical EPEC and EHEC isolates, it was discovered that about half of the strains contain an EspM gene but less than 2% of strains contain EspT (Arbeloa et al., 2009). EspT activates Rac1 and Cdc42 to generate membrane ruffles that results in the eventual phagocytosis of the bacteria (Fig. 2C) (Bulgin et al., 2009a,b). This novel finding of an enteroinvasive EPEC (and Citrobacter) may illustrate an evolutionary divergence and eventual emergence of a new class of pathogens (Bulgin et al., 2009a). The more common effector EspM is similar to the Shigella protein IpgB2 and activates RhoA to induce stress fibres in infected cells (Fig. 2C) (Arbeloa et al., 2008; 2010). During infection, expression of EspM inhibits the formation of the actin pedestals that are characteristic of A/E lesion pathogens (Simovitch et al., 2010). The biochemical and cellular mechanisms attributed to this process have yet to be elucidated, but should provide important insight into A/E lesion pathogenesis. Additional questions remain about EspT and EspM. Specifically, why do some strains have EspT and EspM while others do not and what is the pathogenic impact of these GEFs?
Conclusions and perspectives
The SopE/WxxxE family of bacterial GEF mimics provides an interesting insight into how bacterial pathogens exploit small GTPases. Rather than inhibiting signal transduction through these GTPases, these GEF mimics are responsible for orchestrating GTPase mediated complex cellular behaviours including phagocytosis and membrane trafficking. While these bacterial GEFs are evolutionary distinct from their eukaryotic counterparts, they activate and select GTPases using a similar biochemical strategy. Furthermore, similar to eukaryotic GEFs, growing evidence suggests that bacterial GEF mimics are also regulated by complicated protein and membrane interactions. Future investigations on the dynamic interplay between these regulators, their GEFs and the activation of the GTPases will lend insight into the design principles of these virulence factors. Lastly, investigations comparing and contrasting the bacterial GEF mimics and the eukaryotic GEFs will undoubtedly increase our understanding of the processes that maintain host homeostasis and its disruption during infection.
Acknowledgements
Due to space limitations we were unable to comment on all of the reports describing these bacterial GEFs, and we apologize to our colleagues whose work was not discussed. We would like to thank our colleagues Andrey Selyunin and Bethany Weigele for helpful discussions in preparation of this manuscript. This work was supported NIH grants AI083359 (N.M.A.) and AI007520 (R.C.O.) and the Welch Foundation I-1704 (N.M.A.).