A bacterial pore-forming toxin forms aggregates in cells that resemble those associated with neurodegenerative diseases
Summary
Listeria monocytogenes is a bacterial, facultative intracellular pathogen, which secretes a pore-forming toxin called listeriolysin O (LLO). LLO mediates the dissolution of the phagosomal membrane allowing L. monocytogenes to reach and grow in the host cytosolic compartment. In this study we report the localization of LLO secreted in infected cells. We described that LLO (i) forms small perinuclear aggregates, (ii) accumulates in large autophagosome-like structures and (iii) sequesters to large protein aggregates. The formation of protein aggregates required full LLO activity. Further characterization of protein aggregates indicated that they not only contained the active form of LLO but also polyubiquitinated proteins and p62, which are both common components of protein aggregates found in neurological diseases. Hence, a protein of bacterial origin could potentially follow the same fate as a toxic protein associated with neurodegenerative disease.
Introduction
Listeria monocytogenes is a Gram-positive, facultative intracytosolic, food-borne pathogen of animals, pregnant women and immunocompromised individuals, where it often causes neurological disease (Bartt, 2000; Vazquez-Boland et al., 2001). Its primary virulence factor is a secreted pore-forming cytolysin called listeriolysin O (LLO). LLO is an absolutely essential determinant of pathogenesis whose role is to mediate dissolution of the phagosome that results upon internalization (Schnupf and Portnoy, 2007). LLO is a member of the cholesterol-dependent cytolysin family (CDC). CDCs are secreted as soluble monomers that bind and oligomerize to form a prepore at the surface of cholesterol-rich membranes. Conversion into a pore involves the unfurling of two α-helices into β-hairpins that span the membrane bilayer to form a β-barrel that lines the 25 nm aqueous channel (Tweten, 2005).
There are a number of potential mechanisms that allow LLO activity to be compartmentalized to an acidic phagosome. First, LLO is 10 times more active at acidic pH than at neutral pH (Portnoy et al., 1992), and LLO synthesis is controlled at the translational level (Schnupf et al., 2006a). L. monocytogenes mutants that are altered in this translational regulation become cytotoxic to the infected host cell and are consequently avirulent. Nevertheless, some LLO is secreted into the host cell cytosol and is rapidly phosphorylated, ubiquitinated and degraded by the proteasome. Pharmacological inhibition of the proteasome or LLO mutants with extended half-lives are not cytotoxic, suggesting the presence of other mechanisms that may prevent LLO activity in the host cell cytosol (Schnupf et al., 2007; Schnupf et al., 2006b).
The fate of LLO in infected cells has been studied by genetic and biochemical approaches, but surprisingly, LLO has never been visualized by immunofluorescence in infected cells. In this study, we make the surprising finding that LLO secreted inside of cells is largely associated with large aggregates. Protein aggregation is a hallmark of neurodegenerative diseases such as Alzheimer's, Parkinson's and Huntington's diseases (Ross and Poirier, 2004). These aggregates contain polyubiquitinated proteins and are also rich in p62 (also known as sequestosome1) (Zatloukal et al., 2002), an adapter protein that contains a PB1 domain in its N-terminus and an ubiquitin-associated domain. The N-terminus has the ability to interact with the subunit S5a/Rpn10 of the proteasome whereas the ubiquitin-associated domain non-covalently binds polyubiquitin chains. Owing to its ability to bind both ubiquitin and proteasome, p62 might act as an adapter capable of binding and storing ubiquitinated proteins for subsequent delivery to proteasomes (Wooten et al., 2006). Here we report that LLO aggregates that form in the cytosol of infected cells colocalize with ubiquitin and p62, and thus, resemble aggregates associated with neurological diseases (Zatloukal et al., 2002).
Results
Distribution of LLO in infected BMDMs
Using a monoclonal antibody specific for LLO we performed indirect immunofluorescence microscopy in bone marrow-derived macrophages (BMDMs) infected for 8 h with wild-type (WT) L. monocytogenes. LLO was readily detected as either a punctate signal forming circular structures around the nucleus or condensed in large clusters (Fig. 1A). As expected, no signal was detected in cells infected with the LLO-minus mutant (Fig. 3D). Further analysis of LLO clusters showed that they could be divided into two categories: very large clusters that were either associated with a tight clump of bacteria or big clusters free of bacteria. Immunofluorescence performed at 0.5, 2, 4, 6 and 8 h post infection (p.i.) showed that the above pattern of LLO localization became apparent as soon as 4 h p.i.
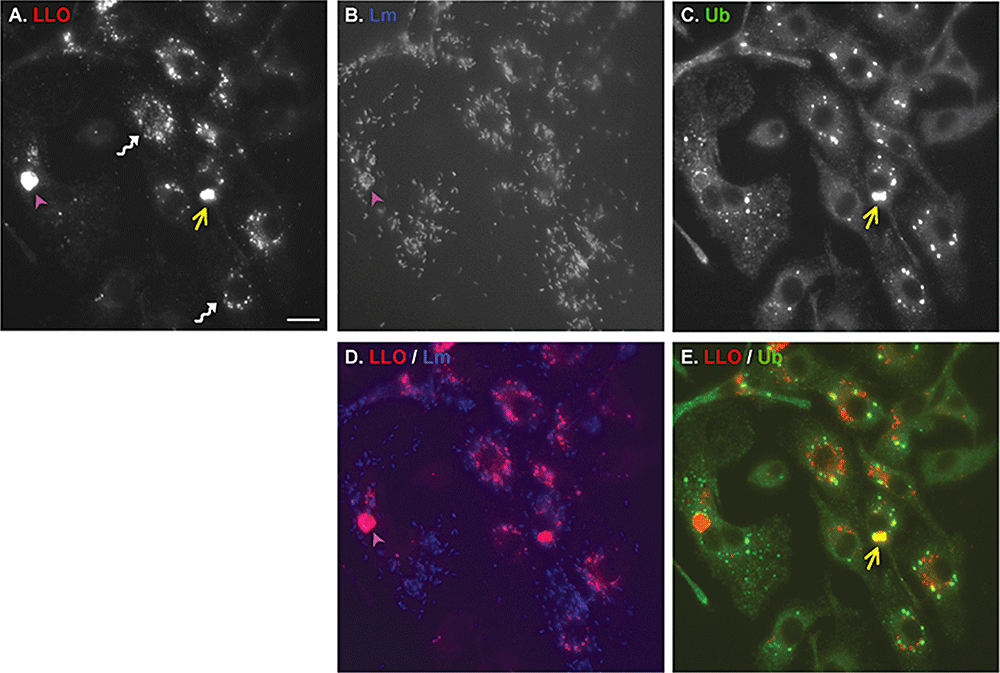
Localization of listeriolysin O in infected cells. Immunofluorescences of BMDMs infected with wild-type L. monocytogenes, 8 h post infection and stained for LLO (A), L. monocytogenes (B) or ubiqutin (Ab FK2) (C). D. Merge of LLO (red) and Listeria (blue) stainings. E. Merge of LLO (red) and ubiquitin (green) stainings. Twisted white arrows indicate small perinuclear aggregates of LLO. Purple arrowheads indicate autophagosome-like structures, i.e. colocalization of LLO with a cluster of L. monocytogenes. Yellow arrows indicate large aggregates of LLO, i.e. colocalization of LLO with ubiquitin aggregate. Autophagosome-like structures did not colocalize with ubiquitin staining although occasionally, a thin thread of ubiquitin highlighted the edge of the structure. Scale bar, 5 μm.
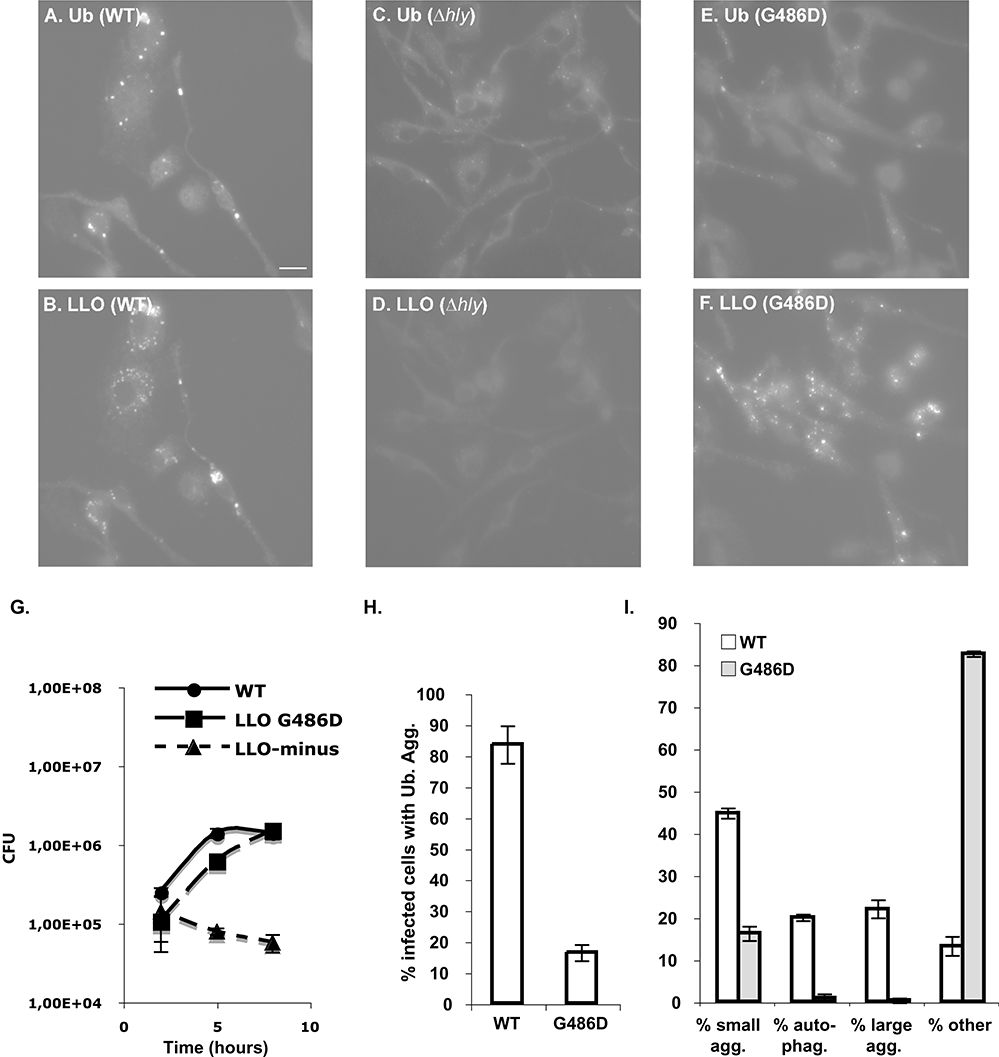
LLO and ubiquitin aggregates require LLO activity. BMDMs were infected with L. monocytogenes wild-type (WT), LLO-minus (Δhly) or LLOG486D-producing strains (G486D). Cells were fixed 8 h p.i.A–F. Immunofluorescence micrographs stained for conjugated ubiquitin (Ab FK2) and LLO. Scale bar, 5 μm.G. Intracellular growth of wild-type, LLOG486D and LLO-minus strains in BMDMs.H. Percentage of infected cells containing ubiquitin aggregates.I. Distribution of LLO. See Experimental procedures for calculations. Agg., aggregates; auto-phag., autophagosome-like structures.
The LLO aggregates associated with a group of bacteria resembled large vacuoles (Fig. 1A, B and D). These vacuoles were on average, 5–9 μm in diameter and morphologically similar to the vacuole-like structures containing group A Streptococcus observed in infected HeLa cells (Nakagawa et al., 2004) or the Francisella-containing vacuoles observed in infected BMDMs (Checroun et al., 2006) that were both positive for the autophagy marker LC3.
The LLO aggregates that were free of bacteria colocalized with ubiquitinated proteins (Fig. 1A, C and E).
For simplicity, we will refer to these three different patterns of LLO localization as small aggregates, autophagosome-like structures and large aggregates. Observation of 100 cells 8 h p.i., in triplicate, showed that LLO was distributed as follow: 45% small aggregates, 22% large ubiquitin aggregates, 20% autophagosome-like structures, and 13% others (Fig. 3I). In this study, we concentrated on the large ubiquitin aggregates.
Formation and characterization of ubiquitin aggregates
Listeriolysin O was found in aggregates containing ubiquitinated proteins. Formation of ubiquitinated protein aggregates in response to L. monocytogenes infection was very frequent, as about 83% of infected cells showed multiple ubiquitin aggregates. These aggregates varied in size with an average of approximately 2 μm in diameter. We sought to compare and contrast the ubiquitin aggregates formed in BMDMs upon L. monocytogenes infection with the several types of intracellular aggregates containing ubiquitinated proteins that have been described in the literature.
Aggresomes are thought to form as a deposition site for proteins that accumulate owing to an overwhelmed proteasome. Aggresomes are microtubule-dependent inclusion bodies that colocalize with markers of the microtubule organization centre (MTOC) such as γ-tubulin (Kopito, 2000; Corboy et al., 2005). Co-staining for ubiquitin and γ-tubulin of BMDMs infected with L. monocytogenes showed that the ubiquitin aggregates did not colocalize with γ-tubulin (Fig. 2A). Furthermore, whereas aggresome formation is blocked by drugs such as nocodazole that depolymerize microtubules, treatment of WT L. monocytogenes-infected BMDMs with nocodazole from 2 to 8 h p.i. did not prevent the appearance of ubiquitin aggregates (Fig. 2B). These results indicated that ubiquitin aggregates triggered by L. monocytogenes infection were not aggresomes.
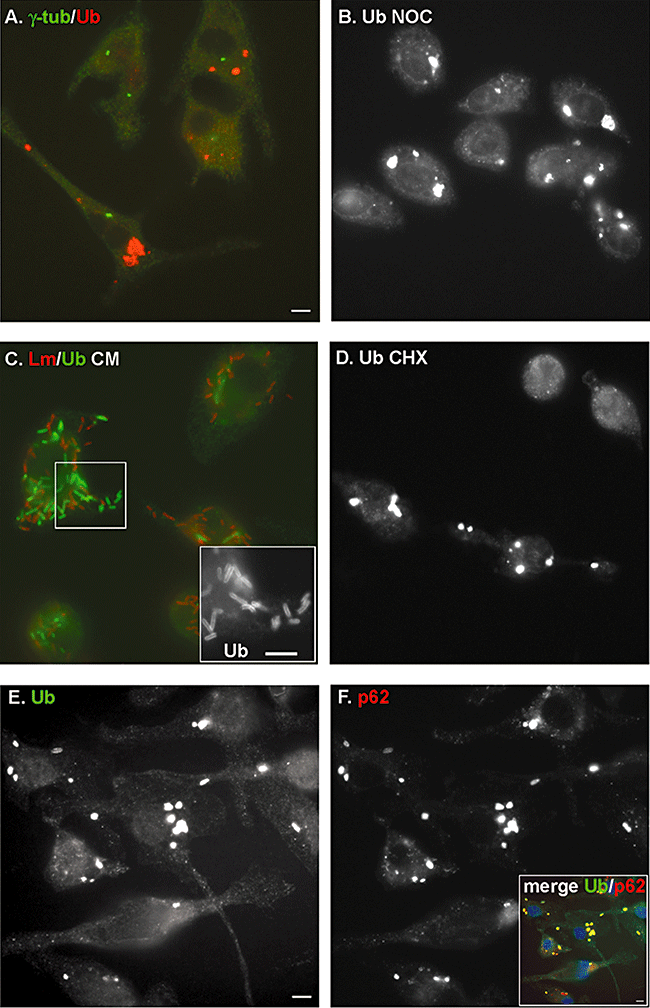
Characterization of ubiquitin aggregates that accumulate during L. monocytogenes infection. In all micrographs BMDMs were infected with wild-type L. monocytogenes.A. Ubiquitin aggregates [stained in red (Ab FK1)] do not colocalize with the MTOC (γ-tubulin, stained in green).B. Staining of ubiquitinated proteins (Ab FK2) shows that the formation of ubiquitin aggregates is not sensitive to the microtubule depolymerizing drug nocodazole (10 μm added from 2 h to 8 h p.i.).C. Ubiquitin aggregates disappear 24 h p.i. after treatment with the bacterial translation inhibitor chloramphenicol (20 μg ml−1 for 16 h). Instead, ubiquitin [stained in green (Ab FK2)] is observed surrounding bacteria (stained in red). Inset is a magnification of ubiquitin staining to highlight the localization of ubiquitin at the bacterial surface.D. Staining of ubiquitinated proteins (Ab FK2) shows that ubiquitin aggregates are not sensitive to the eukaryotic translation inhibitor cycloheximide (20 μg ml−1 added at the time of infection until 8 h p.i.).E–F. Images of ubiquitin (Ab FK2) and p62 stainings, respectively, in infected cells 8 h p.i. Inset shows colocalization (yellow) of p62 (red) and ubiquitinated proteins (green) in aggregates. Cell nuclei were stained with DAPI (blue) in the merge picture. Scale bars, 5 μm.
Another type of protein aggregate, triggered by inflammatory stimuli, are aggresome-like induced structures (ALIS). In contrast to aggresomes, ALIS do not require microtubule transport and do not colocalize with MTOC markers. ALIS, first described in Dendritic cells (and formerly called DALIS), are transient aggregates of ubiquitinated proteins that require continuous protein synthesis (Lelouard et al., 2002). ALIS formation has been reported in macrophages upon exposure to L. monocytogenes (intact bacteria or culture supernatant) (Canadien et al., 2005). In an attempt to determine if formation of ubiquitinated aggregates was dependent on a specific innate immune activation pathway we evaluated the formation of aggregates upon L. monocytogenes infection in BMDMs isolated from MyD88−/−, IRF3−/−, Nod1−/− or Nod2−/− knock-out mice. Neither the absence of MyD88, the common adapter to Toll-like receptors, or the absence of the transcriptional factor IRF3 or the absence of cytosolic receptors Nod1 and Nod2 prevented the formation or morphological appearance of ubiquitin aggregates (data not shown).
Aggresome-like induced structures formation peaks 8 h post stimulus and aggregates disappear after 24–36 h. To test if the ubiquitin aggregates resulting from infection with L. monocytogenes were also transient, we performed immunofluorescence specific for conjugated ubiquitin 24 h p.i. rather than the usual 8 h p.i. During this extended time-course of infection, host cell death was prevented by blocking bacterial growth after 8 h with the translation inhibitor chloramphenicol. As shown in Fig. 2C, the ubiquitin aggregates disappeared after 24 h p.i., and ubiquitin was closely associated with individual bacteria. These results are similar to the observations made by Perrin et al. who described the association of ubiquitin with the L. monocytogenes cell surface in the presence of chloramphenicol (Perrin et al., 2004). The disappearance of ubiquitin aggregates supported the hypothesis that the ubiquitin aggregates that appear during L. monocytogenes infection were ALIS. However, whereas ALIS formation requires continuous host translation, we were not able to prevent their appearance by treating cells with the eukaryotic translation inhibitor cycloheximide (Fig. 2D). However, as noted above, they disappeared in the absence of bacterial protein synthesis. Therefore, those aggregates could be ALIS whose formation would rely on continous bacterial translation instead of host translation, unfortunately the lack of specific marker for ALIS limits further characterization.
The formation of ubiquitin aggregates is a hallmark of degenerative diseases. Those aggregates include Lewy bodies in Parkinson's disease, neurofibrillary tangles in Alzheimer's disease, huntingtin aggregates and Mallory bodies in steatohepatitis. The adapter protein p62 is a common component of these aggregates (Zatloukal et al., 2002; Nagaoka et al., 2004). In order to determine if ubiquitin and therefore LLO aggregates contained p62, we performed immunofluorescence on WT L. monocytogenes-infected BMDMs using antibodies against the C-terminal portion of p62 and conjugated ubiquitin. Strikingly, p62 colocalized with the ubiquitin aggregates (Fig. 2E and F). Non-infected cells contained a low level of p62 aggregates (data not shown) that increased dramatically in number and size upon infection.
The role of p62 during L. monocytogenes infection
Although p62 seems generally required for aggregate formation it has already been reported that downregulation of p62 using small interfering RNA did not affect aggregate formation (Nagaoka et al., 2004). We tried to address the precise role of p62 in the formation and/or removal of aggregates by infecting BMDMs from p62−/− knock out mice with WT L. monocytogenes. However, under these conditions, ubiquitin aggregates were still observed, indicating that p62 was not required to cluster polyubiquitinated proteins (Fig. S1). To ensure that ubiquitin aggregates were still transient, p62−/− and control BMDMs were infected with the WT strain, and the disappearance of ubiquitin aggregates was checked 24 h p.i. and similar results were observed in both population of macrophages (data not shown). Lastly, intracellular growth of WT L. monocytogenes was similar in both population of macrophages (data not shown).
Formation of ubiquitin aggregates requires LLO activity
Aggregates found in neurodegenerative diseases contain, among other things, ubiquitinated proteins, p62 and the toxic form of a protein associated with the disease. As ubiquitin aggregates formed upon infection with L. monocytogenes contained LLO, we asked if their formation was dependent on the presence of active (potentially toxic) LLO. BMDMs were infected with the WT and LLO-minus L. monocytogenes strains. The LLO-minus mutant is unable to escape from the phagosome and therefore is unable to grow. To compensate for the defect in growth and to compare cells heavily infected in both cases, BMDMs were infected with 20 times more LLO-minus bacteria than WT bacteria. Under those conditions, infection with the LLO-minus strain did not trigger formation of ubiquitin aggregates comparable to those induced by WT infection (Fig. 3C). As expected, no signal was detected for LLO (Fig. 3D). The presence of LLO was therefore required to trigger the formation of the ubiquitin aggregates. However, a caveat of this experiment was that cells with bacteria growing in the cytosol were compared with cells harbouring bacteria trapped in phagosomes. To further associate the toxicity of LLO with the formation of ubiquitin aggregates, BMDMs were infected with a strain expressing a non-toxic form of LLO: LLOG486D. This strain harbours a point mutation in LLO that still allows bacteria to escape from a phagosome and to grow in the cytosol, but prevents LLO toxicity because LLOG486D is severely crippled, especially at neutral pH (Decatur and Portnoy, 2000; P. Schnupf and D.A. Portnoy, unpubl. data). Infection with the WT or the LLOG486D-expressing strains allowed recovery of the same number of colony forming unit from BMDMs 8 h p.i. (Fig. 3G). However, the LLOG486D-expressing strain was unable to trigger formation of ubiquitin aggregates comparable to the ones observed during WT infection (Fig. 3E–I). Indeed, the number of cells with aggregates was low (Fig. 3H), these aggregates showed weak fluorescence intensity, and on average, were reduced in size twofold to threefold compared with infection with the WT strain.
Detection of LLO and p62 in aggregate-enriched fraction
As an alternative approach to immunofluorescence, we prepared soluble- and aggregate-enriched fractions from L. monocytogenes-infected BMDMs and performed Western blot analysis for detection of LLO and p62 (Fig. 4).
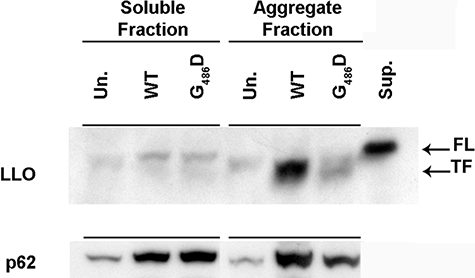
Western blot showing accumulation of the truncated form of LLO and p62 in aggregates-enriched fraction. Immunoblot against LLO (top) and p62 (bottom) present in 1% Triton X-100 soluble and aggregates-enriched fractions (see Experimental procedures) prepared from BMDMs uninfected (Un.) or infected for 8 h with the wild-type (WT) or the LLOG486D-producing strain (G486D). Signal observed for LLO and p62 corresponded to proteins running between the marker of molecular weight 50 and 75 kDa. As a running control for LLO, a supernatant extract from a L. monocytogenes culture broth was loaded (Sup.). FL and TF designate the full length and truncated form of LLO respectively.
First, p62 was preferentially found in the aggregate-enriched fractions from cells infected with WT L. monocytogenes. Second, p62 was already detectable in the soluble fraction and its expression was increased by infection with L. monocytogenes. An increase in p62 mRNA levels by L. monocytogenes infection was also confirmed by microarrays analysis of BMDMs infected with WT or LLO-minus (J.H. Leber and D.A. Portnoy, unpubl. data).
In agreement with the microscopic observations, LLO was also detected in aggregate-enriched fractions when cells were infected with the WT strain but not with the LLOG486D-expressing strain (Fig. 4). Interestingly, the LLO species detected in the aggregate-enriched fraction ran at an apparently lower molecular mass than the LLO secreted by bacteria in culture (Fig. 4). As previously published, two forms of LLO are detected in infected cells, a full-length 58 kDa form and a truncated 55 kDa form. Therefore, it seemed that the LLO found in aggregates is the truncated form of LLO.
Discussion
We report the formation of specific structures in primary macrophages infected with L. monocytogenes. The first structure described, corresponds to a group of bacteria that seem spacially confined in an organelle of 5–9 μm on average, in which LLO is accumulating. Based on the literature, we suggested that these are autophagosome-like structures (Nakagawa et al., 2004; Checroun et al., 2006). We also observed that LLO accumulated in small perinuclear aggregates that are yet to be fully characterized. In this study, we focused on the characterization of the last structure described, which corresponds to aggregates of polyubiquitinated proteins colocalizing with the host protein p62 and the secreted bacterial toxin, LLO. These aggregates vary in size with an average of approximately 2 μm. The formation of the large aggregates was dependent on full LLO activity.
The physiological function of this response to L. monocytogenes infection is not yet understood. Indeed, it is not clear if the LLO aggregates are harmful or beneficial to the host cell. Formation of abnormal protein aggregates is a common feature of degenerative diseases and has been associated with pathogenesis. However, aggregate formation does not necessarily correlate with cell death (Seibenhener et al., 2004; Bjørkøy et al., 2005; Ross and Poirier, 2005). Similarly, in our case, formation of aggregates might provide a mechanism to sequester LLO and prevent damage to cholesterol-containing membranes.
There are a number of clear mechanisms that prevent LLO toxicity including translational control and its pH activity optimum. Further, there are also a number of suspected mechanisms including its phosphorylation, ubiquitylation, proteolysis and short half-life. However, pulse-chase analysis of LLO synthesized in the cytosol revealed two molecular species of LLO, an upper band that was degraded with a half-life of approximately 15 min as determined by the N-end rule pathway of protein degradation, and a stable, slightly truncated form (Schnupf et al., 2006b; Schnupf et al., 2007). Perhaps, LLO that escapes degradation is cleaved by an undefined protease thereby generating a new N-terminus, perhaps one that results in an extended half-life and leads to protein aggregation. Mutation affecting the N-end rule pathway of protein degradation are poorly ubiquitinated and have extended half-lives (Schnupf et al., 2007). These N-end rule mutants show the same pattern of aggregation as WT LLO, indicating that prior ubiquitylation is probably not necessary for LLO to form aggregates (data not shown). However, if the LLO protein present in the aggregates is ubiquitylated, it is possible that ubiquitylation occurs in the aggregate. ALIS, for example, have been described as ubiquitylation centres, as they colocalize with an E1 ubiquitin-activating enzyme, the ubiquitin-conjugating enzyme E225K, as well as the E3 ubiquitin ligase CHIP (Lelouard et al., 2004).
The ultimate fate of the LLO-containing aggregates is not clear. Other than degradation by the proteasome, another possibility is that autophagy contributes to their removal. Perhaps, the entire aggregate is readily enclosed in an autophagosomal structure rather than progressively eliminated through proteasomal degradation. Consistent with this hypothesis, aggregates containing p62 have also been shown to be eliminated by autophagy (Bjørkøy et al., 2005; Pankiv et al., 2007) and impairment of autophagy leads to accumulation of ubiquitin-positive aggregates (Komatsu et al., 2005; Mizushima, 2005). We examined colocalization of LC3 and ubiquitin by immunofluorescence in GFP-LC3 BMDMs and observed that about 50% of ubiquitin aggregates colocalized with LC3 (data not shown). However, this result should be interpreted with caution as two recent publications reported that incorporation of LC3 into protein aggregates was autophagy independent (Szeto et al., 2006; Kuma et al., 2007).
Ubiquitin-positive and p62-positive protein aggregates are a hallmark of degenerative diseases. The aggregate-prone nature of LLO has been observed in vitro. Indeed, it has recently been published that LLO monomers denature in vitro at neutral pH and temperatures above 31°C due to the premature unfolding of transmembranes β-hairpins that normally insert into the membrane to form the β-barrel pore (Schuerch et al., 2005). This is the first report that a bacterial pore-forming toxin follows the same fate as proteins associated with degenerative neurological disease. Interestingly, a comparative analysis has recently highlighted that disease-associated amyloid proteins share in vitro five properties common to bacterial pore-forming toxin: (i) aggregation, (ii) β-sheet formation, (iii) pore formation, (iv) membrane permeabilization, and (v) toxicity (Lashuel and Lansbury, 2006). Therefore, the properties of LLO that lead to its aggregation may provide an interesting model to understand cellular protein aggregation in general. It is also interesting to speculate that LLO aggregates that form in infected cells may be linked to neurological disease caused by L. monocytogenes (Bartt, 2000; Vazquez-Boland et al., 2001).
Experimental procedures
Bacterial strains, mice, cell cultures and infection
Listeria monocytogenes strains used were the WT strain 10403s (Bishop and Hinrichs, 1987), the DP-L2161 strain containing an in-frame deletion of the hly gene (LLO-minus) (Jones and Portnoy, 1994) and the DP-L4044 strain containing a point substitution G486D in LLO (LLOG486D) (Decatur and Portnoy, 2000). Bacteria were grown overnight in brain–heart infusion broth at 30°C without shaking. Before infection, bacterial cultures were washed and re-suspended in equal volumes of PBS.
Bone marrow-derived macrophages were prepared from femurs from C57BL/6 mice (Jackson Laboratory), from femurs from GFP-LC3 transgenic mice obtained from M. Swanson, Department of Microbiology and Immunology, University of Michigan Medical School, Ann Arbor, Michigan; or from femurs from knockout mice obtained from the following sources: myd88−/− from R. Medzhitov, Yale University School of medicine, New Haven, Connecticut; irf3−/− from G. Cheng, Department of Microbiology, Immunology and Molecular Genetics, University of California. Los Angeles, California; nod1−/− from Millennium, Cambridge, Massachusetts; nod2−/− (Mariathasan et al., 2006) from V. Dixit, Genentech, South San Francisco, California; p62−/− from J. Moscat, Genome Research Institute, University of Cincinnati, Cincinnati, Ohio. BMDMs were cultured in DMEM (Gibco) containing 20% heat-inactivated FBS (Hyclone) and 10% 3T3-cell conditioned medium.
Bone marrow-derived macrophages were infected with L. monocytogenes strains at an moi of 10–15:1 for 30 min. The medium was then replaced and 50 μg ml−1 gentamicin was added 1 h p.i.
Inhibitors and antibodies
For pharmacological treatments, infected cells were incubated with nocodazole 10 μm (Sigma-Aldrich), cycloheximide 20 μg ml−1 (Sigma-Aldrich) or chloramphenicol 20 μg ml−1 added 8 h p.i. (Sigma-Aldrich). For immunofluorescence, the following primary antibodies were used: mouse monoclonal IgM anti-polyubiquitinated proteins FK1 and IgG1 anti-mono and polyubiquitinated proteins FK2 (Biomol, 1/1000), mouse monoclonal IgG1 anti-γ-tubulin (Sigma, 1/1000), guinea pig polyclonal anti-p62 C-ter specific (Progen Biotechnik, 1/250), mouse monoclonal IgG1 anti-GFP (Roche, 1/200), rabbit polyclonal anti-Listeria (Difco, 1/1000), mouse monoclonal IgG2b anti-LLO B3-19 obtained from tissue culture supernatants of hybridoma cells (a kind gift of P. Cossart, 1/250). Secondary antibodies isotype specific and conjugated to Alexa-488 (green), Alexa-568 (red) or Alexa-350 (blue) were purchased from Molecular Probes (1/1000). For Western blot, rabbit polyclonal anti-LLO (DP-L2813, 1/10000) and guinea pig polyclonal anti-p62 C-ter specific (Progen Biotechnik, 1/1000) were used. Anti-rabbit and guinea pig Ig coupled to horseradish peroxidase were used as secondary antibodies.
Immunofluorescence
Glass coverslips with infected cells were washed once in PBS, fixed in −20°C cold methanol 10 min at −20°C and washed twice in PBS. Blocking (30 min at 37°C) and antibody incubations (1 h at room temperature) were performed in PBS, 0.1% Triton X-100, 2% BSA. Each incubation was followed by four washes in TBS-T buffer (25 mM Tris-HCl pH 8, 150 mM NaCl, 0.1% Triton X-100). Coverslips were mounted with Vectashield mounting medium (Vector laboratories). Samples were observed at ×600 or ×1000 with a NIKON Eclipse TE 300 inverted microscope. Images were acquired with Metamorph and processed with ImageJ and Photoshop. All colocalizations were checked by confocal microscopy (Zeiss LSM 510 Meta UV/Vis confocal microscope).
For LLO distribution, the number of small aggregates, autophagosome-like structures and large aggregates was counted on 100 cells showing a signal for LLO, then presence of ubiquitin aggregates was checked in the corresponding infected cell and this was done in triplicate for each condition. The size of ubiquitin aggregates was analysed on 10 different fields (at least 50 ubiquitin aggregates) using metamorph program.
Aggregates enrichment and Western blot
Aggregate enrichment was performed as described by Lelouard et al. (2004). Briefly, cells were collected using ice cold PBS Mg2+ and Ca2+ free, and submitted to a 30 min extraction in 20 mM Tris-HCl pH 7.4, 1.5 mM MgCl2, 10 mM NaCl, 1% Triton X-100 at 4°C, followed by a 30 min DNase I (10 units, Invitrogen) treatment at 37°C in the same buffer and a 30 min incubation at 4°C in 20 mM Tris-HCl pH 7.4, 1.5 mM MgCl2, 2 M NaCl. Pellet was washed and re-suspended in PBS.
About 40 μg of proteins of a 1% Triton X-100 extracted fraction and aggregates-enriched fraction was loaded on NuPAGE 10% Bis-Tris Gel, run in MOPS buffer (Invitrogen), and proteins were transferred to a nitrocellulose membrane. Immunoblots were revealed by chemiluminescent ECL advance or ECL + detection kit (Amersham).
Acknowledgements
We thank Maria Diaz-Meco, Jorge Moscat, Michele Swanson for providing us with femurs from knockout and transgenic mice. We thank members of the Portnoy lab for help with BMDMs preparation and helpful discussion.
We thank Jennifer Robbins and the Drubin lab members for help with microscopy.
We thank Pamela Schnupf and Tâm Mignot for fruitful discussion and critical review of the manuscript.
The research was supported by the National Institutes of Health AI27655 (to D.A.P.). D.A.P consults and has a financial interest in Cerus Corporation, a company that might stand to benefit from the results of this research.




