How Developments in Cryobiology, Reproductive Technologies and Conservation Genomics Could Shape Gene Banking Strategies for (Farm) Animals
Contents
Many local breeds are currently at risk because of replacement by a limited number of specialized commercial breeds. Concurrently, for many breeds, allelic diversity within breeds declines because of inbreeding. Gene banking of germplasm may serve to secure the breeds and the alleles for any future use, for instance to recover a lost breed, to address new breeding goals, to support breeding schemes in small populations to minimize inbreeding, and for conservation genetics and genomics research. Developments in cryobiology and reproductive technology have generated several possibilities for preserving germplasm in farm animals. Furthermore, in some mammalian and bird species, gene banking of material is difficult or impossible, requiring development of new alternative methods or improvement of existing methods. Depending on the species, there are interesting possibilities or research developments in the use of epididymal spermatozoa, oocytes and embryos, ovarian and testicular tissue, primordial germ cells, and somatic cells for the conservation of genetic diversity in farm- and other animal species. Rapid developments in genomics research also provide new opportunities to optimize conservation and sampling strategies and to characterize genome-wide genetic variation. With regard to gene banks for farm animals, collaboration between European countries is being developed through a number of organizations, aimed at sharing knowledge and expertise between national programmes. It would be useful to explore further collaboration between countries, within the framework of a European gene banking strategy that should minimize costs of conservation and maximize opportunities for exploitation and sustainable use of genetic diversity.
Introduction
Globally, genetic diversity in farm animals is under threat. Many local breeds are currently at risk (Rischkowsky and Pilling 2007) as they became largely displaced because of the global use of a limited number of specialized commercial breeds. Some local breeds may be at risk because they become very rare, whereas other local ‘heritage’ breeds are not numerically scarce, but at threat because of their very localized regional distribution. Breeds may be lost altogether, but also allelic variation within breeds may decline because of genetic drift or high inbreeding rates within populations.
Local breeds are seen as ‘cultural heritage’, and they may fulfil specific requirements with respect to local terrain or climate, or product quality of typical regional products (Gandini and Villa 2003). Perhaps more importantly, they may carry traits or alleles that may be very useful in specific conditions or for future breeding goals. Also, with respect to the more widely used breeds, it is necessary to preserve within-breed genetic diversity. The commercial breeds are represented by large numbers of animals, but the genetic diversity of these breeds may in fact be quite small (Stachowicz et al. 2011), as a small number of sires are selected to have a multitude of progeny, leading to high rates of inbreeding. Apart from the problems associated with inbreeding, a loss of genetic diversity reduces the genetic ‘toolbox’ necessary for continued breeding. Therefore, prevention of further loss of breeds and of genetic diversity within local and commercial breeds should have top priority.
One strategy to curb further loss of genetic diversity is to somehow ensure continued use of (rare) breeds, by in situ conservation or conservation by utilization. This can be achieved, for example, by subsidizing farmers who maintain animals of rare or local breeds, but this is not always considered as a sustainable and politically acceptable option for the long term. The preferred approach is to improve the self-sustainability of the breeds by promoting factors that ensure that the local breeds are continued to be used in the future (Hiemstra et al. 2010).
Despite such efforts, many native breeds will only be maintained in small and local populations. Local populations may be threatened by calamities such as large scale culling measures in the case of outbreak of infectious diseases. In addition, both rare and commercial breeds run the risk of continued loss of alleles. Therefore, in situ conservation needs to be complemented by gene banking to secure the germplasm of rare and commercial breeds (Woelders et al. 2004; FAO 2007). Long-term storage of material is used to preserve the genetic diversity of today for any use in the (more distant) future, for instance to re-establish a lost breed and to restore genetic diversity within a breed to healthy levels, or to use specific genotypes to address new breeding goals. Secondly, gene bank resources can be, and in fact are, actively used to support breeding schemes of rare breeds to minimize inbreeding (Sonesson et al. 2002; Woelders et al. 2004). This requires a regular update of material in the gene bank and a ready availability of gene bank resources for use in the field. Another purpose of gene bank resources is to make available material for research, for instance to genetically characterize breeds and describe their phylogeny.
Types of Germplasm and Tissue Types to be conserved
Developments in cryobiology and reproductive technology have generated several possibilities for preserving germplasm in farm animals, for example, semen, embryos, oocytes, ovarian and testicular tissue, primordial germ cells (PGCs), stem cells, somatic cells to be used in cloning, and freeze-dried germplasm. Feasibility and cost-efficiency of the collection, freezing and use of these types of germplasm may differ among species and for various situations or local conditions. In addition, feasibility and cost-efficiency depend on the purpose of the gene bank, that is, on whether the germplasm is likely to be used regularly or not.
Semen
For many animal species, there are adequate protocols for collection and freezing of semen, and for its use in artificial insemination (AI), and for some (farm) animal species, there is an existing ‘infrastructure’ for the collection and use of semen in AI. Semen seems very well suited for supporting breeding schemes of small breeds, especially in those species (e.g. cattle, horse) where AI is practised anyway and the use of the frozen semen does not incur extra costs. Semen can also be used to reconstruct a lost breed, but it will take 4–7 generations of backcrossing to restore the ‘original’ genotype, depending on the desired purity of the restored breed. This is time-consuming and costly, and it means that a large number of breeding doses need to be stored in the gene bank. The required number of doses is smaller if sexed semen is conserved, so that in each generation, predominantly female offspring is obtained that can in turn be inseminated with the sexed cryopreserved semen. Perhaps, this may only be possible in situations in which the sexing technology is (commercially) available, that is, in the case of semen of regular AI bulls. Another disadvantage of semen is that mitochondrial genes are not conserved, as sperm mitochondria do not contribute to the offspring’s mitochondrial population.
Embryos
Embryos are also used in gene banks for the conservation of genetic diversity, as they allow fast recovery of a lost breed in a single generation. Consequently, also fewer embryos than semen doses need to be collected, frozen and stored to allow recovery of a lost breed. Boettcher et al. (2005) showed that combined storage of semen and embryos could be most (cost)effective. Yet, easy and cost-effective use of embryos in gene banks may be limited to these species for which collection and transfer techniques are available and operational. This is certainly the case for cattle (Van Wagtendonk de Leeuw et al. 1997; Youngs 2011) and to a lesser extent also for sheep (Youngs 2011). Live offspring or pregnancies have also been reported for other farm animal species, including pigs (Li et al. 2009), horse (Ulrich and Nowshari 2002; Galli et al. 2007), goat (Rodríguez-Dorta et al. 2007) and rabbit (Naik et al. 2005), but in most of these species, the techniques are not routine.
Oocytes
Similar to embryos, restoration of a lost breed or genotype by using cryopreserved oocytes (plus semen) would not require backcrossing. In recent years, both slow freeze and vitrification protocols render excellent results in humans resulting in many healthy babies born (Porcu and Venturoli 2006; Cobo and Diaz 2011). Less data are available in animal species (see, e.g., Critser et al. 1997) than in humans. Live-born young produced from cryopreserved oocytes have been reported in cattle (Abe et al. 2005), mouse (Stachecki et al. 2002) and horse (Maclellan et al. 2002). The collection of oocytes through ovum pickup (ovum aspiration) procedures can be relatively efficient and could be a way to harvest germplasm from animals that are not capable or are not available for producing offspring or embryos. Yet, for many species, techniques of in vitro maturation, cryopreservation and IVF or ICSI of oocytes are currently not available or inefficient.
Ovaries
Cryopreservation of ovaries could be another way to conserve the genotype of animals. In the human, ovary banking can be used to prevent loss of fertility during cancer treatment of women. Cryopreserved ovaries or parts of ovaries may be used as a source of oocytes, or oocytes may be harvested from heterotopically grafted ovaries to be subsequently used in an IVF procedure to produce embryos (Terazono et al. 2012). In animals, an alternative would be to graft cryopreserved ovarian tissue or whole ovaries orthotopically in a recipient animal to restore fertility in that animal. This animal can then be mated and will produce offspring carrying the ovary donor genotype (see some examples in mice and in birds explained later). A similar approach is transplantation of cryopreserved testicular tissue (Song and Silversides 2007).
Stem cells, primordial germ cells and somatic cells
In theory, spermatogonial stem cells can be harvested from a recipient animal and lead to sperm production in a donor male (Izadyar et al. 2002, 2003; Trefil et al. 2010). Furthermore, this is not likely to be an easy and cost-efficient way to conserve genetic diversity. As explained in the following section, in birds, cryopreservation of PGCs may be an efficient way to preserve genotypes, breeds or breeding lines.
Somatic cells can be used to produce embryos, as was first demonstrated by the creation of the sheep ‘Dolly’. Live offspring have been obtained from cloned embryos in a large number of mammalian species, including the farm animal species cattle, sheep, goats, buffalo, pigs, horse and mule (Campbell et al. 2007). Several companies offer cloning as a commercial service. Collection of suitable somatic cells is straightforward. Cryopreservation protocols for somatic cells are relatively simple and do not require controlled-rate freezing equipment. This means that establishing the collection is easy and cheap. The opposite is true, however, for using the material when needed. Utilization involves culturing the cells after thawing, reprogramming of the nuclei, collection of oocytes by ovum pickup or from slaughtered animals, culture and in vitro maturation of the oocytes, enucleation of the oocytes, transfer of the somatic nucleus to an enucleated oocyte, culture of the resulting embryos and finally, embryo transfer (ET) in recipients of the same or a related species. In this procedure, the donor mitochondrial genes are not conserved. Because of the low upfront costs, a gene bank of somatic cells was established as a backup for endangered livestock breeds in conditions with little infrastructure in Vietnam (Groeneveld et al. 2008).
Farm animal gene bank collections in the Netherlands
Apart from minor collections of embryos and oocytes, the Dutch gene bank of the Centre for Genetic Resources, the Netherlands (CGN) currently contains relatively large collections of semen (Table 1). CGN covers only farm animal species and species or breeds within species that may have a farm-related history. In addition to the rare native breeds, CGN also preserves regular updates (‘snapshots’) of semen from the breeding bulls and boars of the Dutch commercial cattle and pig breeding organizations. The gene bank has an EU-certified storage site for cattle semen, and semen stocks of different species and of different veterinary health status are stored separately. In addition, for security reasons, we have duplicate collections in two separate locations. CGN resources are used in the field to a limited extent to support breeding schemes of rare breeds (mainly cattle) in particular to keep inbreeding levels at an acceptable level. Perhaps, for those cattle breeds, a minimum of 100 breeding units per male are retained as ‘core collection’ for long-term conservation of allelic variation. The aim is further to maintain stocks of at least 25 males per breed in long-term storage for any future use, for example, reconstruction of a lost breed.
| Species | No. breeds | No. males/breed | No. straws |
|---|---|---|---|
| Bovine | 9 | 7–4095 | 181 753 |
| Dog | 2 | 3–7 | 162 |
| Goat | 2 | 6–25 | 3820 |
| Horse | 5 | 8–20 | 18 200 |
| Pig | 16 | 7–47 | 69 981 |
| Chicken | 20 | 5–19 | 18 827 |
| Sheep | 7 | 12–68 | 22 147 |
Specific Problems and Potential Solutions
Whereas there are many potential types of germplasm that can be used in repositories for animal genetic resources, there are a number of species and situations within species in which gene banking of material is difficult, impossible, or very costly, requiring development of new alternative methods or improvement of existing methods.
Semen
Semen is a versatile and often low cost option for gene banking of (farm) animal genetic diversity. Yet, for a number of species, sperm freezing methods give unsatisfactory results and further method development is required. In some other species, not the freezing per se, but rather the collection of semen may be the problem. In cattle, one obstacle is that no or not enough bulls of rare breeds are offered by AI stations and semen from bulls must be collected on farm, which is officially prohibited under EU veterinary regulations. In very small and inbred populations, there is a strong possibility that the semen quality of many or all of the remaining males is very low, and so cryobanking may be even more limited. In particular situations, collection of semen may be too costly or fails altogether. For instance in more or less feral animals, like the sheep of various heath breeds, the males may be too ‘wild’ to be trained for the collection of semen. We found that the ability to train rams for semen collection varied strongly between breeds. Therefore, CGN and other groups have tried the collection and use of epididymal spermatozoa for sheep (Woelders et al. 2004, 2005; Ehling et al. 2006). In sheep, the yield of spermatozoa from the caudae epididymidis is very high, well above 20 billion spermatozoa per animal in mature rams, allowing the production of more than 100 insemination doses (200 million sperm/dose) per animal. In addition, the freezability (post-thaw per cent motile and live sperm) and fertilizing ability of frozen-thawed epididymal spermatozoa from Veluwe rams (a Dutch heath sheep breed) appeared to be at least as good as that of ejaculated semen from rams of the same breed (Table 2). In the Netherlands, since 2004, epididymal spermatozoa have been used predominantly to produce the gene bank collections for Dutch native sheep breeds (Table 3). Also, in breeds where ejaculated semen can be collected, we found that collection of epididymal spermatozoa is most cost-efficient. The samples can be used without requiring special facilities or expertise, and no housing and training of the rams is required. We collaborate with the breed organizations and are notified when genetically suitable rams are being culled after having served for natural inseminations in the herd, and testes are collected and transported from the abbattoir to the institute, to be processed the same day. The total costs involved are very limited in comparison with the collection of ejaculates to the same number of doses (Woelders and Kaal 2008). We use a simple rapid method of obtaining the spermatozoa in a semi-quantitative way (Woelders et al. 2005; Woelders and Kaal 2008), which involves cutting the cauda (submersed in Tris egg yolk freezing medium) with a scalpel many times so it becomes more or less minced and squashed (see, e.g., Soler et al. 2003). This allows virtually all spermatozoa to escape from the convoluted epididymal duct. Alternative methods are float-up or retrograde flushing (e.g. Cary et al. 2004; Martinez-Pastor et al. 2006; Santiago-Moreno et al. 2009).
| Ejaculated semen (5 rams) | Epididymal spermatozoa (3 rams) | |||
|---|---|---|---|---|
| Motile sperm (%) | 42 ± 4.5 | 60 ± 0 | ||
| Live sperm (%) | 48.8 ± 2.1 | 62.3 ± 5.6 | ||
| Pregnant | Lambs/ewe | Pregnant | Lambs/ewe | |
| Cervical AI | 0/11 | 4/10 | 2.0 | |
| Laparoscopic AI | 6/10 | 2.3 | 7/10 | 3.1 |
- AI, artificial insemination.
| Breed | Rams ejaculate | Rams epididymal | Rams total | Doses/breed | Mean doses/ram |
|---|---|---|---|---|---|
| Mergelland | 21 | 4 | 25 | 3038 | 122 |
| Kempen heath sheep | 18 | 15 | 33 | 5048 | 153 |
| Veluwe heath sheep | 7 | 28 | 35 | 3790 | 108 |
| Schoonebeeker | 10 | 15 | 25 | 2905 | 116 |
| Drente heath sheep | 2 | 58 | 60 | 4794 | 80 |
| Melkschaap | 30 | 1 | 31 | 1521 | 49 |
| Zwartbles | 7 | 5 | 12 | 1078 | 90 |
For similar practical reasons, epididymal spermatozoa are used for the conservation of genetic diversity of rare wildlife species, for example, the European bison (Kozdrowski et al. 2011), Red deer (Martinez-Pastor et al. 2006) and Ibex (Santiago-Moreno et al. 2009).
There are few publications regarding collection of epididymal spermatozoa in avian species (Clulow and Jones 1988; Kwon et al. 1995; Orlu and Egbunike 2010). The morphology and function of the epididymis in birds are quite different than in mammals. In quail, it was shown that most ‘extragonadal’ sperm are found in the vas deferens (Clulow and Jones 1988) rather than in the epididymal duct. In chicken, interesting sperm numbers can be retrieved from the vasa deferentia (2–4 billion sperm per male, C. Zuidberg, CGN, personal communication), and experiments are currently underway at CGN to investigate collection, freezability and post-thaw fertilizing ability of vas deferens sperm in birds in the interest of finding efficient ways of collection and cryopreserving Dutch native breeds of ducks, geese, chicken and pigeon.
Sheep AI
In sheep and goats, AI with fresh semen is possible and is being used in practice. Methods for freezing ram or buck semen are available and give a fair rate of success (Salamon and Maxwell 2000; Leboeuf et al. 2008). With frozen-thawed ram semen, good fertility results are obtained with laparoscopic insemination (surgical semen deposition in the uterine horns), but not with ‘normal’ cervical insemination. The reason seems to be that frozen-thawed spermatozoa have a shorter lifespan and probably a less efficient motility, and therefore, sperm migration through the cervix and uterus to reach the site of fertilization is impaired. Non-surgical intrauterine insemination is not easy with conventional techniques and instrumentation, because of the complex anatomy of the cervix. Laparoscopic insemination is prohibited in the Netherlands, and in any case, it is more complicated and time-consuming than conventional AI. Therefore, development of a non-surgical method of cervical passage would be desired for ewes. Similarly, intrauterine insemination could also be beneficial in goats.
The anatomy of the sheep cervix is highly complex, which makes it difficult to pass the cervix with an implement (Halbert et al. 1990a; Kershaw et al. 2005). Three approaches have been tried to reduce the difficulty of passing through the cervix. One is by using an implement to retract the cervix into the vagina to identify the cervical opening and align the funnel-shaped rings of the cervix to some degree (Halbert et al. 1990a,b). A second approach is the use of an agent to relax the cervix (e.g. PGE-2 or PGE analogues, oxytocin, interleukin-8, misoprostol, or hyaluronan (see several references in Wulster-Radcliffe et al. 2004; Perry et al. 2010), or the beta-adrenergic blocking agent carazolol (Gündüz et al. 2010). A third approach is designing appropriate transcervical AI and ET equipment to overcome the physical difficulties associated with the ovine cervix (Wulster-Radcliffe et al. 2004). In what is referred to as the Guelph system, restraint of the ewe and instrumentation were adapted to allow transcervical deposition of the semen in at least a fraction of all tested ewes (Halbert et al. 1990b), which resulted in higher pregnancy rates compared with vaginal semen deposition in the vagina or cervical os. In later studies, both improved transcervical AI implement (Wulster-Radcliffe et al. 2004) and administration of PGE2 (Candappa et al. 2009) or PGE1 analogue (Leethongdee et al. 2007) allowed 100% penetration of the cervix, but lambing data are either missing or not dramatically improved. In the case of goats, excellent kidding rates were reported after transcervical AI with frozen-thawed semen (Sohnrey and Holtz 2005). Furthermore, it appears that in sheep, further research is needed.
Horse
The problems and potential solutions associated with cryobanking of horse germplasm are reviewed in full detail by Smits et al. (2012) and will be discussed here only briefly. In the horse, recovery of a lost breed using semen is much more expensive than in other farm animal species (Gandini et al. 2007). This may be improved when using low dose (1–10 million sperm) insemination techniques, for example, by hysteroscopic insemination (Morris and Allen 2002). At the same time, however, the possibilities of using embryos or oocytes appear to be poor in the horse. Cryopreservation of horse embryos appears to be very difficult indeed (Tharasanit et al. 2005), with relatively few reported pregnancies (see references in, e.g., Choi et al. 2011) and only a few live births reported after transfer of cryopreserved embryos (Ulrich and Nowshari 2002; Galli et al. 2007). This is an area where horse reproductive specialists and cryobiologists should work together to improve the methods for cryopreservation of horse embryos taking into account the special morphological and other embryo characteristics seen in this species (see, e.g., recent advances in Choi et al. 2011; Scherzer et al. 2008).
Collection of horse oocytes is in itself a practical possibility, as oocytes can be collected, without knowing the stage of the estrous cycle, by transvaginal ultrasound-guided ovum pickup, allowing the collection of more than 70 oocytes per year per mare. But cryopreservation of horse oocytes is very difficult (Tharasanit et al. 2006a,b), with only one report of live offspring after transfer of embryos produced from cryopreserved oocytes (Maclellan et al. 2002).
Although the horse has the disadvantage in the field of embryos and oocytes compared with some of the other farm animal species, it seems to have an advantage with regard to the production of offspring from embryos generated from (cryopreserved) somatic cells by nuclear transfer, with many healthy live offspring and high efficiencies reported. Possibilities and successes are reviewed by Smits et al. 2012 and in the FAO Draft Guidelines for the Cryoconservation of Animal Genetic Resources (FAO 2011). In the Netherlands, CGN has considered cryobanking of somatic cells as a means to preserve the genotypes of unique, old mares for which other approaches to preserve their germplasm are currently not possible or impractical. The ethical acceptability of nuclear transfer ‘cloning’ is, however, debated, partly because of concerns of animal health and welfare, as cloning in many species is still associated with higher levels of embryonic loss and malformed young (Campbell et al. 2007; Oback 2008), but also because of other ethical considerations. One could still consider cryobanking of somatic cells in case no other viable route is available, on the premise that the material will not be used unless future methods regarding nuclear reprogramming have evolved to a stage that offspring can be obtained without compromising health or welfare or the genetic makeup in the first offspring and in subsequent generations. An overview of possible routes and areas for future research are shown (Fig. 1).
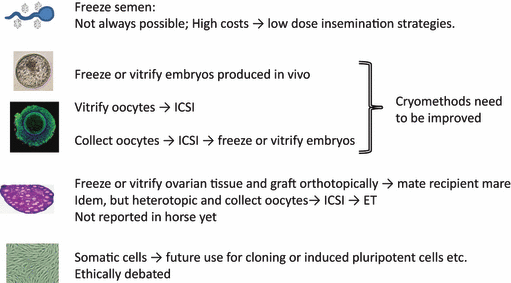
An overview of potential routes and areas for future research for cryobanking of horse germplasm
Laboratory animals
At the turn of the millennium, more than 2500 mutant mouse lines existed (European-Commission 2001), and since then, the number of genetically modified mouse lines has rapidly expanded, and ambitions are to produce a comprehensive resource of mouse mutants in which each of the approximately 20 000 genes in the mouse genome has been knocked out (NIH 2006). Institutions and core facilities offering mouse archiving/retrieving services all offer embryo and approximately half offer sperm cryopreservation as well. Few institutions provide ovary cryopreservation and recovery services. A feasible cryopreservation strategy should enable reconstitution of the original genotype, including the congenic background, in one or two generations. Embryo banking is an option, but in mutant mouse lines, poor response to induced superovulation and poor oocyte or embryo quality may be encountered. Both the mutation and the genetic background can impair embryo development (Glenister and Thornton 2000). Regardless, the costs involved in embryo banking are high (Glenister and Thornton 2000; European-Commission 2001; Landel 2005), several thousands of Euros for the cryopreservation of a single strain.
Semen banking may seem an obvious choice, as cryopreservation of mouse semen is easy and quick. If the semen can be used to fertilize oocytes of females of the same inbred background, only one generation breeding with the F1 offspring would be sufficient to recover the homozygous mutant against the original genetic background. Yet, the same problem here is that in the inbred strains, IVF results may be insufficient (Glenister and Thornton 2000).
The cryopreservation and use of ovaries for conserving a mutant genotype at first glance seems a less obvious option, as success may depend on fine surgical skills and requires good post-thaw survival of the ovaries that are able to revascularize, and elicit no graft rejection, etc. Yet, recent progress in cryopreservation and use of mouse ovaries has shown that this approach has great promise and may have several advantages over the banking of semen and embryos. Cryopreservation methods seem to have improved and are in fact made simpler by using vitrification protocols very similar to those used for embryos and oocytes (Huang et al. 2008, 2010). More importantly, it was shown that grafting is a relatively simple and successful procedure, which was demonstrated by using cryopreserved ovaries of a large series (22) of mutant strains with various genetic backgrounds (C57BL/6, FVB, BALB/c) to recover the genotype by orthotopic grafting of the vitrified ovaries in recipient mice (Huang et al. 2010). The method appears to be relatively easy and remarkably efficient. Collection of ovaries and subsequent vitrification of the ovaries are quick and simple, taking approximately 2 h per session. It requires no hormonal treatments of the females. The vitrified ovaries can be orthotopically transplanted in a simple and quick surgical procedure. Again, no preparation of the recipient females is required. Using recipients of the same genetic background as the donors prevents graft rejection. Within 10–14 days of transplantation, recipients can be bred to congenic wild-type males to obtain live offspring that is heterozygous for the mutation of interest, but homozygous for genetic background, with a success rate of >40% offspring of the donor genotype. For recovery of the mutant strain, only 5–10 ovary recipients and 1–2 males (both with proper wild-type isogenetic background) are needed to reconstitute the strain. The total cost of archiving and retrieving a mutant mouse line by ovary tissue freezing may be cheaper than with semen or embryo freezing, considering the number of animals required, and labour and equipment cost (Huang et al. 2008).
Birds
A reliable and efficient strategy for gene banking germplasm of birds would be important for gene banking of rare species of wild birds and of rare breeds of poultry, but would also be important as an alternative to holding and propagating live birds to maintain breeding lines in research institutes and poultry breeding organizations. In birds and other macrolecithal taxa, methods for cryopreservation of the ova or embryos are not available. Cryopreservation of semen is possible in a number of bird species, but may not be possible or efficient in specific cases. For the chicken, adequate methods for freezing semen are available with good fertility being reported in a number of studies using cryopreserved semen (Tselutin et al. 1999; Woelders et al. 2006). CGN recently created a semen gene bank collection of native Dutch chicken breeds and currently has a stock of semen from 20 breeds with 5–19 males per breed (Table 1). Furthermore, validation of the semen freezing method we use at CGN (Woelders et al. 2006) was performed only using semen from a commercial chicken line, and post-thaw fertility of frozen-thawed semen of the rare breeds may be lower than that of the commercial breed. Variation between breeds was in fact reported by Blesbois et al. (2007). In turkey, a recent study also indicated large differences in success of cryobanking of semen for various breeding lines, with no success (no hatched poults) in one of the lines studied (J. A. Long, H. Phillip, P. A. Purdy, C. A. Zuidberg, S. J. Hiemstra, S. G. Velleman, H. Woelders, unpublished data).
Potential alternative types of germplasm for gene banking in bird species are ovaries and PGCs. Recent studies suggested that cryobanking of ovaries is a viable and efficient option in birds. Live offspring carrying the donor genotype was obtained after transplantation of cryopreserved half-ovaries in quail (Liu et al. 2010). Both slow freeze and vitrification protocols were used in that study, but vitrification gave best result. Like in the mouse, the procedures used for collection, vitrification and grafting of the ovaries appear to be remarkably easy to perform and successful. Graft rejection was prevented by oral administration of an immunosuppressant. This method would be ideal if it can be complemented with cryopreserved semen from the same breed, so that the original breed or breeding line can be recovered in a single generation. In that respect, it is interesting that a similar approach of grafting cryopreserved testicular tissue from roosters rendered males that produced (donor testis derived) sperm sufficient for successful intramagmal insemination of hens (Song and Silversides 2007). If no semen of the breed or line is available, several generations of backcrossing would be needed and this would imply that larger stocks of ovaries need to be banked, as would be the case when only semen banking would be used to secure a breed or line.
Another possibility in birds is cryobanking of PGCs. Primordial germ cells can be simply collected from blood from day 3, Hamburger Hamilton (HH) stage 14–16 chicken embryos. (Naito et al. 1999; Van de Lavoir et al. 2006; Macdonald et al. 2010). The PGCs can be cultured and propagated as clonal cell lines and frozen. Frozen-thawed PGCs can then be used to produce chimaeric embryos by injection of the PGCs into the cardiac tract of HH stage 14–16 embryos, which then are transferred into phase III host shells and cultured to hatching in a specialized procedure (Perry 1988). Primordial germ cells only seem to give germline transmission when transferred to a same sex embryo. Male hatchlings can be raised to sexual maturity and may then carry the donor genotype in a fraction (2–16%; Macdonald et al. 2010) of their sperm cells. Likewise, female PGCs in female embryos can generate female chimaeras with the donor genotype in a fraction (0–12%; Van de Lavoir et al. 2006) of the produced ova. Currently, more research is needed to make the procedures more easy and consistent, but in principle, the cryobanking of PGCs would have the advantage of allowing recovery of a lost breed or breeding line without the need of backcrossing, provided that also female PGCs can be cloned and propagated (c.f. Van de Lavoir et al. 2006; Macdonald et al. 2010).
Conservation Genomics
Gene bank material is stored to be useful for several purposes: for example, to support populations to prevent or overcome genetic problems (drift, inbreeding, genetic defects), to reconstruct breeds in case of extinction or loss of substantial numbers of animals, to create new lines/breeds, to quickly modify or reorient selection of breeds, or for research purposes (Woelders et al. 2004). For all these purposes, it is essential that the genetic diversity conserved in the gene bank material is as large as possible and that the whole range of genetic variance is represented. This applies both to the genetic diversity within and across breeds. From a genetic and economic point of view, sampling strategies in cryopreservation programmes should above all maximize genetic diversity in the gene bank and avoid duplications. The number of animals per breed that need to be sampled to cover the genetic variation in a breed depends on the genetic diversity and relatedness within the breed. For instance, if a breed is highly inbred, only few animals need to be sampled to cover all genetic diversity. For the reconstruction of a lost breed, the number of males and the number of breeding units per male also depend on the acceptable level of inbreeding in the reconstruction phase and the required purity of the reconstructed breed, as well as insemination success and litter size. For example, for cattle breeds, as a ‘rule of thumb’, we aim to store at least 200 breeding units per male of at least 25 males per breed.
Characterization of farm animal genetic diversity can be performed with pedigree information and with molecular marker information. Genetic diversity estimated with pedigree information is based on the statistical relation between kinship and diversity, where kinship is defined as the probability that two alleles drawn at random from any gene are identical by descent (copies of the same ancestral allele). Consequently, a high kinship implies low genetic diversity. Pedigree kinships have been used in several studies to estimate genetic diversity within breeds (e.g. Melka and Schenkel 2010; Selvaggi et al. 2010). Pedigree kinship is an accurate estimate for overall genetic diversity, provided that a reliable pedigree is available. Accuracy decreases with low pedigree depth, pedigree errors and missing pedigree data. Missing pedigree data and pedigree errors can result in low estimated kinships even in a population that is highly inbred, resulting in a negative effect on conservation of genetic diversity (Oliehoek et al. 2006; Mucha and Windig 2009). Generally, there are no pedigree data available that characterizes the genetic relationships between breed.
Besides pedigree information, also molecular information can be used to characterize genetic diversity. Initially, only few molecular DNA markers were available, such as blood groups or allozymes. Over the years, an increasing number of variable markers have been developed (Schlotterer 2004). Currently, microsatellites, which are highly polymorphic and distributed all over the genome, are the most widely used markers in genetic diversity estimation of animal populations (e.g. Freeman et al. 2006; Tapio et al. 2010). Perhaps, single nucleotide polymorphisms (SNP) markers are now rapidly gaining importance. Single nucleotide polymorphisms markers are available in large numbers across the genome. Currently, 50 000-SNP chips are widely used in cattle studies and 800 000-SNP chips have become commercially available, whereas generally only 30–100 microsatellites are used in cattle. Single nucleotide polymorphisms markers are now the markers of choice in QTL analysis and genomic selection. In addition, SNP data are now being used for genetic diversity estimation in livestock breeds (e.g. Flury et al. 2010; Lin et al. 2010). Sequencing, that is, typing every single base-pair on the whole genome, is now also available on a commercial basis for several species, but still a costly and complex operation, and not developed for routine determination of genetic diversity.
The availability of SNP markers provides new possibilities for genetic diversity estimation. In contrast to other markers and pedigree-based genetic diversity, SNP markers enable the evaluation of genetic diversity both across the whole genome and at specific parts of the genome. Engelsma et al. (2011) have recently investigated the consequences of using SNP-estimated diversity for gene banks. Methods of choice to maximize genetic diversity in a gene bank are the optimal contribution method (Meuwissen 1997) and the core set method (Eding and Meuwissen 2001) for within-breed and between-breed diversity, respectively. With these methods, contributions of animals or breeds are chosen so that the total diversity measured as (1 – the average kinship) is maximized. In other words, duplication of genetic diversity is avoided while total diversity is maximized. Kinships for these methods can be based on both pedigrees and DNA markers. With the arrival of SNP markers, better estimation of kinships with DNA markers is possible.
Generally, breeds considered for storage in gene banks have reduced population size and consist of highly related animals. Molecular comparisons of genetic diversity between breeds generally require sets of unrelated animals within breeds and breeds that have diverged for a long time. In a recent study in Holstein cattle, Engelsma et al. (2012) demonstrated that with SNP chips, genetic diversity can be compared even for recently diverged groups of highly related animals. Furthermore, with SNP chips, the distribution of genetic diversity over the genome (i.e. the frequency of polymorphisms in chromosomes or chromosome segments) can be analysed as well, something not possible with pedigree analysis.
With the same data, (Engelsma 2012) showed that SNP-based optimal contributions resulted not only in considerably more conserved genetic diversity than random selection of animals, but also in slightly more conserved genetic diversity than with pedigree-based optimal contributions. Also, de Cara et al. (2011) concluded that optimal contribution is more effective with SNP- than with pedigree-based kinships.
In some situations, it is preferred to maintain alleles of a specific gene at predetermined frequencies, for example, if a disease-related allele needs to be minimally represented in the gene bank or if an allele characteristic of a breed needs to be maximally represented. Such a focus on a single gene can negatively affect overall genetic diversity in the gene bank, especially if representation of an allele in the gene bank needs to be largely different from the population frequency, for example, if a common allele needs to be eliminated, and if individuals that contain unique diversity are excluded from the gene bank because they do not have the required allele. Engelsma (2012) demonstrated that use of SNP markers provides the possibility to maximize genetic diversity of the gene bank collection while at the same time focussing on a single gene.
In conclusion, when reliable and complete pedigrees are available, existing methods to maximize genetic diversity generally work well. SNPs provide additional opportunities to conserve genetic diversity without pedigrees or at specific parts of the genome. To optimize sampling strategies from a genetic and economic perspective, developments in conservation genomics should be incorporated in cryopreservation programmes.
European Collaboration of Gene Banks for Farm Animal Genetic Resources
As of 2002, the Centre for Genetic Resources, the Netherlands (CGN) of Wageningen University and Research Centre receives government funding for various tasks related to conservation of genetic diversity of farm animals, including gene banking activities. Other European countries also started cryopreservation programmes. The European Regional Focal Point for Animal Genetic Resources network (ERFP) plays an important role in exchanging experiences and good practices between countries. Recently, the ERFP established a Working Group on Ex Situ Conservation that will promote exchange of knowledge and experiences with gene banking and explore further development of a joint European Farm Animal Gene Bank strategy or framework. Furthermore, the European Commission (AGRI GENRES 870/04 programme) recently funded two projects (HERITAGESHEEP, EURECA) that included substantial cryopreservation components. The fact that breeds may be found in European regions that exceed national borders and the fact that considerable gene pools of a breed may be available in other countries than the country where the breed originated calls for further development of a joint European strategy for gene banking. Perhaps, the most important first step is to increase collaboration with regard to characterization and documentation and to share knowledge and expertise between national programmes, with regard to storage and use of germplasm and of conservation genetics. In specific cases, joint gene banking may be undertaken to optimally use expertise and facilities available and prevent duplication.
Conclusion
Access to new technologies is important for gene banks to develop their collections and to add value to collections. Rapid developments in reproductive technologies, cryobiology and genomics pose new challenges and opportunities for gene banks. At the same time, gene banks need to economize their operations. From this perspective, international collaboration in scientific research, characterization, establishment and maintenance of gene bank collections becomes increasingly important.
Acknowledgements
This work is funded by the Ministry of Economic Affairs and Agriculture in the progamme WOT 436 Genetische Bronnen.
Conflicts of interest
None of the authors have any conflicts of interest to declare.




