Microbial colonisation of artificial and deep-sea sediments in the Arctic Ocean
Abstract
Although environmental factors such as grain size and organic carbon content may influence the distribution of microbes in marine sediments, there has been little experimental study of the topic to date. To investigate how those sediment variables affect microbial colonisation under in situ conditions, deep-sea sediments and artificial sediments (glass beads, sands) were incubated in the Arctic deep sea at 2500 m water depth with or without chitin, one of the most important carbon polymers in marine environments. Microbial abundance, biomass, chitobiase activity and changes in community structure were monitored after 7 days and 1 year. In control sediments without chitin addition, no significant changes in microbial abundance, biomass and activity were observed after 1 year. In the presence of chitin, however, considerable increases in these parameters were recorded in all three sediment types tested. Regardless of chitin addition, natural deep-sea sediments were always associated with higher values of microbial abundance, biomass and activity compared with artificial sediments. Sediment type was always found to be the most significant factor explaining variation in enzymatic activity and bacterial community structure as compared to the effects of chitin amount, incubation time, and changes in cell number or biomass. Overall, this is the first in situ study that has addressed the effects of multiple factors and their interactions on abundance, biomass, activity and community structure of microbial communities in the deep Arctic Ocean.
Problem
Several field experiments in the deep sea have been conducted to study the patterns of colonisation and succession of macro- and meiobenthic organisms (e.g.Levin & Smith 1984; Grassle & Morse-Porteous 1987; Kitazato 1995; Levin et al. 2006). Yet, in situ studies targeting microbial colonisation in deep-ocean environments are scarce, probably for logistical reasons. A number of in situ studies in the Central Indian Ocean Basin using a benthic hydraulic disturber (Deep-Sea Sediment Resuspension System; DSSRS) have directly examined the impact of deep-sea mining on microbial communities by comparing bacterial abundances and activities in pre- and post-disturbed sediments (Nair et al. 2000; Raghukumar et al. 2001). However, these experiments did not examine the effects of various sediment characteristics (e.g. particle size, particle shape) on changes in structure or composition of microbial communities. Noticeably, most studies dealing with microbial colonisation processes in sediments have been performed using time-course experiments in the laboratory with artificial (Yamamoto & Lopez 1985) or abiotic sediments (Findlay et al. 1992), and have mainly focused on estuarine and intertidal microbial communities. The available studies overall indicated that sediment characteristics such as grain size, grain shape and organic carbon content of sediments are important factors influencing the microbial colonisation of sediments (e.g.Nickels et al. 1981). For example, the microbial abundance in marine sediments is directly controlled by sediment surface area and inversely correlated with sediment grade (Llobet-Brossa et al. 1998). A positive relationship has also been found between bacterial abundance and organic carbon content in sediments (Köster et al. 2005).
Despite high chitin concentrations in oceanic ecosystems, surprisingly few ecological studies have examined the effect of large chitin supply on microbial communities as main agents in mineralisation of this degradation-resistant organic matter (Deming & Baross 1993). Chitin is probably the most important biopolymer in marine environments (Gooday 1990), and Cauchie (2002) estimated an annual chitin amount of about 109 tons in marine ecosystems that is exclusively produced by crustaceans. The Arctic Ocean is characterised by large stocks of copepods (Hirche 1997) and high input of chitinous material at the deep sea floor may originate from mass aggregation of planktonic copepods over the sea floor (Hirche et al. 2006) and the sedimentation of their exoskeletons and casings of faecal pellets.
In the present study we investigated for the first time the effects of sediment type on growth, enzymatic activity and composition of deep-sea microbial communities for a short (7 days) and long (1 year) period directly at the Arctic Ocean deep sea floor. Besides fine-grained deep-sea sediments, artificial sediments made of glass beads and sand were also selected to depict simple model sediment particles within the size range representative for medium and coarse sediments. Chitin was then added to the different sediment types to examine the functional responses of microbial communities to large inputs of particulate organic material. Our main hypotheses were that (i) sediment type, ranging from fine-grained and organic carbon-rich sediments to coarse and organic carbon-poor sediments, will impact microbial growth, activity and community structure after a long-term incubation of 1 year under deep Arctic Ocean conditions, and (ii) the nature of available sediments, e.g. particle size, particle shape, organic carbon content, will modulate those microbial responses.
Material and Methods
Experimental design
In situ experiments were carried out in the eastern Fram Strait, west off Spitsbergen (79°5′N, 4°2′E), more precisely at the central station (2500 m) along a depth transect from 1200 to 5500 m water depth that was established in the deep-sea long-term observatory HAUSGARTEN (Fig. 1; Soltwedel et al. 2005). The central HAUSGARTEN station at 2500 m water depth served as an experimental area for our study that used a free-falling benthic lander designed by the Institut Français de Recherche pour l’Exploitation de la Mer (IFREMER) to deploy samples in situ. The benthic lander consists of an aluminium framework that supports four identical round trays, each of which was equipped with four separate round chambers (0.02 m2 per chamber). Two trays with three chambers each were used for the experiments (Fig. 2), whereas the remaining trays were deployed for different purposes. The benthic lander was equipped with a remotely controlled mechanism consisting of spring-loaded discs to open and close the chambers during the passage to and from the deep sea floor to the surface. Each chamber was covered by a grid (5 mm mesh size) to prevent interference from larger animals. Because of space limitation when using benthic landers, the overall experiment reported here was carried out only once.
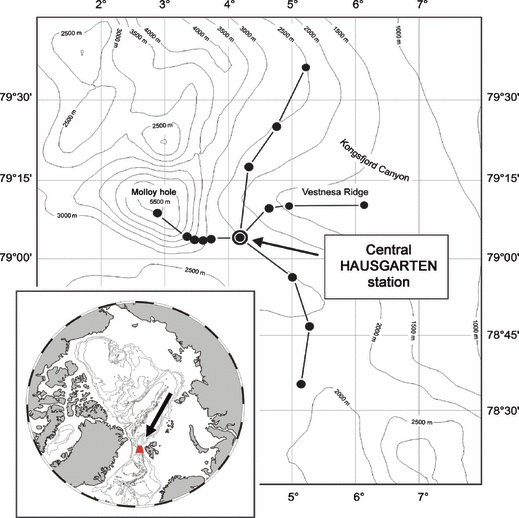
Map of the deep-sea long-term observatory HAUSGARTEN in the Eastern Fram Strait, west off Spitsbergen. Black dots indicate permanent sampling sites along a depth and a latitudinal transect crossing the central HAUSGARTEN station at 2500 m water depth.
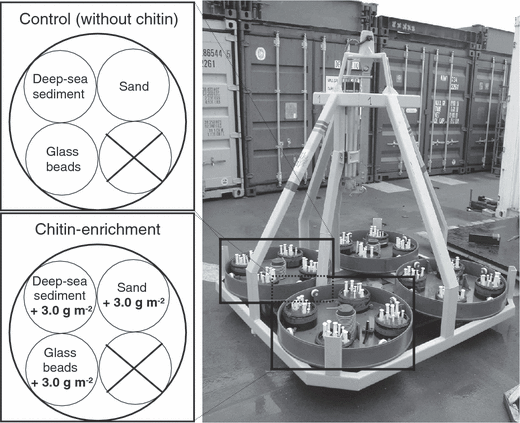
Free-falling benthic lander for in situ enrichment experiments. The lander consisted of four identical trays, each of which was equipped with four separate round chambers. Two trays with three chambers each were used for the experiments (one for the chitin experiment and one for the control without any additional substrate).
Prior to lander deployment, deep-sea sediments from the central HAUSGARTEN station were collected with a multiple corer (MUC; Barnett et al. 1984) at the same depth and close to the same position where the benthic lander ultimately was deployed. One chamber of each tray was filled to 1 cm from its top with surface sediments (0–3 cm) pooled from several sediment cores. The second chamber was filled with glass microbeads (diameter 200 μm; MHG Strahlanlagen GmbH, Düsseldorf, Germany) consisting of simple smooth and spherical particles. The third chamber contained commercially available axenic coarse sand that was selected on the basis of sub-rounded to sub-angular particles with larger mean grain sizes (1–2 mm). Grain sizes in deep-sea sediment were analysed using a Coulter counter. The pore size of artificial sediment composed of glass spheres was calculated as described by Sharma & McInerney (1994). The organic carbon content of the different sediment types was determined by using an Elemental Analyser (EuroVector, Milan, Italy).
Following defaunation by freezing sediment samples at −30 °C for 48 h and subsequent thawing, samples were taken for initial analyses of microbial parameters (0 day). A chitin concentration of 3 g·m−2 chitin (purified chitin flakes from crab shells; Sigma-Aldrich, Hamburg, Germany) was then mixed thoroughly with the upper sediment layers and placed in the lander chambers of the first tray. The addition of chitin was equivalent to a substrate input of 1.3 g organic C m−2 and corresponds to the organic C supply that arrives on the deep sea floor at the HAUSGARTEN central station annually (E. Bauerfeind, personal communication). Chambers of the second tray were filled only with natural and artificial sediments without any addition of organic substrate and served as a control. During the RV Polarstern cruise ARK XX/1 in July 2004, the benthic lander was deployed for 7 days. After recovery and subsequent sampling, trays were equipped with new chambers which were filled with the three defaunated sediment types. Deep-sea sediments for the long-term incubation were taken from MUC sampling for the short-term experiment and were stored frozen until deployment. Amendment of natural and artificial sediments with chitin was carried out in the same way as described above. Afterwards, the benthic lander was deployed at the same station for 1 year. Sampling of the 1-year experiment was carried out during the expedition ARK XXI/1b of RV Polarstern in July 2005.
In addition to microbial parameters (see below), some abiotic parameters were also determined to assess the environmental conditions during the experiments. The bottom water temperature during both expeditions, for example, was about −0.8 °C and bottom water oxygen concentration was 280–320 μmol·l−1. Mean bottom currents were relatively weak, ranging from 5.3 to 6.6 cm·s−1 at 0.1 and 0.9 m above the bottom, respectively (E. Sauter, unpublished data).
Sub-sampling and sample processing
After recovery of the trays, eight samples from the uppermost sediment layers of each chamber were taken by means of plastic syringes with cut-off ends (50 ml, 2.5 cm in diameter). Sub-sampling was carried out in a cold room at 4 °C. The first sediment centimetre from all eight syringes was pooled and homogenised before determining total microbial cell number, enzymatic activity, and community structure.
Microbial biomass
For the determination of microbial cell numbers and biomass, 2 ml of the sediment sub-sample was taken by means of plastic syringes with cut-off ends (5 ml, 1.2 cm in diameter) and stored refrigerated in 9 ml of 2% formaldehyde solution. Total cell numbers were determined by epifluorescence microscopy after staining with acridine orange according to the method of Meyer-Reil (1983). Volumetric measurements of the microbial cells were conducted with the New Porton grid, measuring randomly 50 cells per filter as described by Grossmann & Reichardt (1991). Microbial biomass was estimated using a conversion factor of 3 × 10−13 g C μm−3 (Børsheim et al. 1990). For each sample, 40 counted grids from two replicate filters were analysed.
Enzyme activity
The extracellular enzymatic activity of chitobiase was measured fluorometrically (F-2000 spectrofluorometer; Hitachi) using the methylumbelliferone (MUF) labelled substrate N-acetyl-β-glucosaminide (Sigma-Aldrich) according to Boetius & Lochte (1994). Previous trials to determine the substrate saturation level gave a final concentration of 100 μmol MUF-substrate. Relative fluorescence units were calibrated with seven MUF standard concentrations between 10 and 300 nmol. Enzyme activity was calculated per volume of sediment and time (nmol·h−1) by linear regression. The regression coefficient was always > 0.95. For each sample three measurements per incubation were performed.
Terminal-restriction fragment length polymorphism (T-RFLP) analysis
Sediment samples for microbial community analysis were stored at −30 °C until analysis. Total DNA was extracted and purified using a FastDNA® SPIN Kit for soil (Qbiogene, Heidelberg, Germany) following the manufacturer’s instructions. DNA concentrations obtained from 400 mg sediment ranged from 50 to 200 ng·μl−1. Bacterial 16S rRNA genes were amplified using universal primers 27F and 1492R (both synthesised by Interactiva; Ulm, Germany). The bacterial specific forward primer 27F was end-labelled with phosphoramidite fluorochrome 5-carboxyfluorescein (5′6-FAM). The PCR cocktail contained 2 μl of template DNA, 0.25 μmol (each) primer, 1.5 mmol KCl, 10 mmol Tris–HCl pH 9, 1.5 mmol MgCl2, 250 μmol dNTP, and 1.25 U Taq-Polymerase (Amersham Pharmacia Biotech, Freiburg, Germany) in a final volume of 50 μl. DNA amplification was performed using the following cycling conditions: a 3-min hot start at 95 °C, followed by 29 cycles consisting of denaturation (1 min at 95 °C), annealing (1 min at 55 °C), and extension (1 min at 72 °C). A final extension at 72 °C was then done for 7 min. Fluorescently labelled PCR products were run onto 1% agarose gel and purified using QIAquick Gel Extraction kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions.
Approximately 300 ng of the PCR product was digested with 10 U of endonucleases HhaI and MspI (Amersham Pharmacia Biotech). The digestions were performed separately in a total volume of 50 μl at 37 °C for 5 h. Following desalting by isopropanol precipitation, fragment separation was performed by Gene Analysis Service GmbH (Berlin, Germany) using an ABI Prism 310 capillary analyser (PE Applied Biosystems, Foster City, CA, USA).
The size of end-labelled terminal restriction fragments (T-RFs), visualised as peaks on the resulting electrophoretic profiles, was determined by comparison with an internal size standard (GS2500 TAMRA, PE Applied Biosystems). Peaks between 50- and 1000-bp long were analysed using GENESCAN analytic software 2.02 (ABI). An additional check for artefacts was manually performed and peaks whose areas were smaller than 1% of the total peak area were excluded from data analysis. T-RFLP analyses were done once for each treatment due to the limited amount of sediment samples.
Data analysis
The variables total microbial cell number, biomass, chitobiase activity and organic carbon content were log10-transformed prior to performing statistical analyses to normalise their distribution. Three-way analyses of variance (ANOVA) were applied to test the effects of each factor (sediment type, chitin addition, incubation time) and of their interactions on each dataset. Differences between treatment means of total microbial cell numbers, biomass, chitobiase activity and organic carbon contents were evaluated by using pairwise Tukey’s honestly significant difference (HSD) test at P < 0.05 following significant ANOVA tests.
For each sample, T-RFLP fingerprints obtained from the two different restriction digests were combined to generate a binary matrix according to the presence or absence of T-RFs. The unweighted pair group with arithmetic mean (UPGMA) method was applied with the Jaccard similarity index. Nodal support in the resulting dendrogram was determined by performing 100 bootstrap replicates (Hammer et al. 2001). In addition, non-metric multidimensional scaling (nMDS; Schiffman & Reynolds 1981) was used to obtain an ordination of samples based on a matrix of Jaccard dissimilarities of T-RFLP profiles. Twenty iterations of the nMDS procedure based on different random initial positions of the samples were performed so as to obtain an ordination with the lowest stress value (i.e. the best goodness-of-fit). Various groupings among samples were depicted on the ordination and tested for significance using analysis of similarities (ANOSIM; Clarke 1993) tests based on 1000 permutations.
A distance-based redundancy analysis (db-RDA; Legendre & Anderson 1999) was used to determine how well different factors explain the variation in bacterial community structure. The overall procedure consists of the following steps: A dissimilarity matrix among samples is first calculated by using the Jaccard coefficient, and then principal coordinate analysis is applied to the dissimilarity matrix to obtain new, uncorrelated coordinates which were then analysed as independent variables in redundancy analysis (Legendre & Legendre 1998; Ramette 2007). To assess the respective importance of the different factors and of their covariation on community structure, a variation partitioning procedure (Borcard et al. 1992; Ramette & Tiedje 2007) was subsequently applied to the data. This technique assesses the respective effects of each factor after controlling for the effects of other factors using a combination of simple and partial redundancy analyses. Significances of the respective effects of each factor were tested using 1000 Monte Carlo permutation tests. Variation partitioning analysis was also used to assess the respective contribution of each factor and of factor covariation to changes in chitobiase activity. Statistical analyses were performed with the statistical software JMP IN version 5.1 (SAS Institute, Cary, NC, USA) and with the VEGAN package (R version 2.4.0; The R foundation for statistical computing).
Results
Effects of sediment type, chitin enrichment and incubation time on microbial abundance and activity
In deep-sea sediments from the central HAUSGARTEN station, silt (4–63 μm) was the dominant grain-size fraction (43%), followed by sand (≥ 63 μm, 37%), and clay (< 4 μm, 20%). In contrast to this natural fine-grained sediment, coarse sand and spherical glass beads were provided as artificial sediments for microbial organisms to colonise. Three-way analyses of variance revealed that each factor taken alone (sediment type, chitin input, and incubation time) had highly significant effects on microbial cell number, biomass and chitobiase activity (Table 1). Moreover, the analyses showed that factor interactions also explained some of the variation of the measured variables. Microbial cell number was, however, the only exception, as a non-significant F ratio was obtained when the interactions among the three factors were evaluated. The interactions between chitin and incubation time explained the largest variation of all pairwise interaction terms, although sediment type and incubation time displayed the largest variation when each factor was considered separately (Table 1).
| factorsa | microbial cell number | microbial biomass | chitobiase activity | |||
|---|---|---|---|---|---|---|
| dfb | F ratioc | df | F ratio | df | F ratio | |
| sediment type | 1 | 288.3*** | 1 | 246.8*** | 1 | 402.3*** |
| chitin | 2 | 102.56*** | 2 | 652.1*** | 2 | 91.3*** |
| time | 2 | 262.2*** | 2 | 775.3*** | 2 | 170.0*** |
| sediment type × chitin | 2 | 5.5* | 2 | 8.5** | 2 | 7.4** |
| sediment type × time | 4 | 6.4** | 4 | 428.7*** | 4 | 54.7*** |
| chitin × time | 2 | 43.0*** | 2 | 13.2*** | 2 | 35.5*** |
| sediment type × chitin × time | 4 | 1.2n.s. | 4 | 8.4*** | 4 | 5.5** |
- aThree-way ANOVAs were performed on log10-transformed variables. Factors consisted of chitin input (presence or absence), sediment types (deep-sea sediment, glass beads, sand) and incubation times (0, 7 days, 1 year). Statistical differences between treatment means are indicated in Fig. 3.
- bDegrees of freedom (df) for each factor or interactions thereof.
- cF ratios are ratios of the mean–square value for a given source of variation to the residual mean–square value. Associated probabilities are indicated as not significant (n.s.) when P ≥ 0.05, (*) when P < 0.05, (**) when P < 0.01, (***) when P < 0.001.
More detailed analyses of the variation in microbial cell number (Fig. 3a,b), microbial biomass (Fig. 3c,d) and chitobiase activity (Fig. 3e,f) indicated overall very similar patterns, i.e. generally higher values for deep-sea sediment than for glass or sand substrates and a remarkable increase in chitin-enriched samples after a 1-year incubation. In the no-chitin, control treatments, initial microbial cell numbers were significantly higher in natural sediments from the study site (0.6 × 108 cells·cm−3), than in glass beads (0.1 × 108 cells·cm−3) and sands (0.2 × 108 cells·cm−3; Fig. 3a,b). Microbial cell numbers present in the artificial sediments at the start of the experiment were not different from background contamination due to sample processing. When chitin was added to the experimental system, the only significant changes in cell number occurred after 1 year; no noticeable differences were found for shorter incubation times regardless of sediment type (Fig. 3b). Natural sediments were then clearly associated with the highest cell number followed by sand and glass beads.
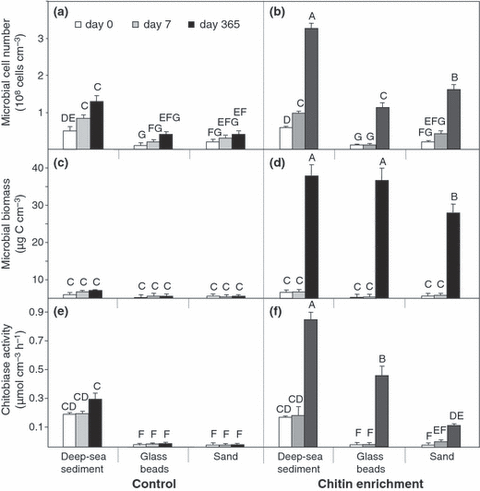
Mean microbial cell number, biomass and chitobiase activity in control (a,c,e) and chitin-enriched (b,d,f) sediments after different incubation times. The concentration of 3.0 g chitin m−2 added to the sediments corresponds to 1.3 g C·m−2. Capitalised letters (A–G) indicate significant differences between mean treatments as determined by post hoc Tukey’s HSD tests at P < 0.05. Significance letters above bars can only be meaningfully compared between panels (a,b), (c,d), and (e,f).
Changes in microbial biomass in the different treatments also confirmed the trends observed for microbial cell number (Fig. 3c,d). Indeed, in treatments without chitin, no changes were observed over time, whereas chitin addition was associated with higher microbial biomass, but only after 1 year of incubation. However, in contrast to cell numbers, no significant difference between deep-sea sediment and glass beads was observed in microbial biomass after 1 year. Interestingly, whereas changes in microbial biomass were mainly accompanied by increased cell numbers in chitin-enriched, deep-sea sediment, the increased microbial biomass for artificial sediments was mainly associated with a substantial increase in the mean cell volume and less with the increase in cell number (Fig. 3b,d). With the glass beads used in our experiment, the calculated pore size of 17 μm was much larger than an average bacterial cell and thus adequate for the colonisation by larger bacterial rods; the latter was not detected by microscopic observations in natural sediments or in control sediments without chitin (data not shown).
Measurements of extracellular enzymatic chitobiase activity clearly showed differences between treatments (Table 1) consistent with interactions of sediment type, chitin input and incubation time. Sediment type had the largest effect on chitobiase activity (Fig. 3e,f). Initial chitobiase activity from deep-sea sediment of about 0.2 μmol·h−1 corresponded to that of the natural environment and to values estimated for other deep-sea sediments (Boetius & Lochte 1994, 1996; Boetius 1995). A large increase in enzymatic activity was detected only after 1-year incubation (Fig. 3f), which clearly suggests that the addition of chitin induced the production of chitobiase. Furthermore, there was also a significant relationship between decreasing particle size (from sand, glass to deep-sea sediment, respectively) and increasing chitobiase activity.
Because the effects of incubation time, sediment type and chitin addition were tightly intertwined, further analyses were needed to disentangle the respective effects of each factor and to quantify their covariation (Fig. 4a,b). Sediment type was found to be the most important factor (18%, P < 0.001), explaining variation in microbial activity when the effects of incubation time, chitin addition and concomitant changes in cell number were controlled (Fig. 4a). The relative effects of chitin input and incubation time, although significant, each accounted for only 6% of the variation in enzymatic activity (Fig. 4a). Interestingly, there was also a high covariation between cell number and both sediment type and incubation time, accounting for 16% and 18%, respectively, of the variation in chitobiase activity. When microbial biomass was used instead of microbial cell number as a factor (Fig. 4b), a larger proportion of the overall variation in microbial activity (70%) could be explained (Fig. 4b) compared with 60% for cell number (Fig. 4a). The largest amount of the variation in activity could then be explained by the presence of different sediment types and by fluctuations in microbial biomass, as well as the covariation between those factors (Fig. 4b). Noticeably, neither pure effects of chitin nor incubation time were found significant when sediment type and microbial biomass were considered (Fig. 4b).
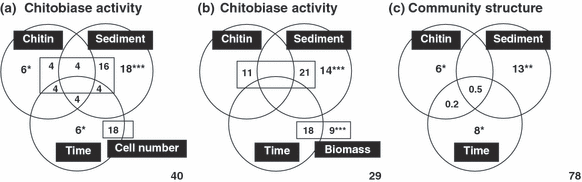
Variation partitioning analysis as a function of sediment type, chitin input, incubation time, microbial abundance and biomass. The respective contribution (as percentages of the total biological variation) of each factor and of their covariation was disentangled by using variation partitioning and distance-based redundancy analyses. (a) Variation in chitobiase activity was partitioned into the respective effects of chitin input, sediment type, incubation time and total cell number. (b) Variation in chitobiase activity was partitioned as for the (a) panel but with total microbial biomass replacing microbial cell number. Percentages within the square inserted into the circles represent covariation between microbial growth parameter (cell number, biomass) and the effects of chitin input, sediment type and incubation time. (c) Variation in community structure as determined by T-RFLP was partitioned in the effects of the four factors. The respective effects of total cell number and microbial biomass as well as their covariation with other factors did not contribute significantly to the community variation (all below 5%) and are not displayed. The significance of each fraction was tested by using 1000 permutations of the data (* P ≤ 0.05, ** P < 0.01, *** P < 0.001). Negative variations were not shown, and the amount of unexplained variation is indicated at the bottom right hand corner of each panel.
Finally, the organic carbon content of deep-sea sediment we used was about 0.9% by weight at the start of the experiment and did not significantly change after short- and long-term incubation (Table 2). In the three-way ANOVA that tested the effects of incubation time, chitin input and sediment type on organic carbon content, the only non-significant term was that of the incubation time by chitin interactions (data not shown). Effects of each single factor and factor interactions were highly significant (P < 0.01), and the overall interactions between incubation time, chitin input, and sediment type were marginally significant (F-ratio = 3.403, P = 0.046). Glass beads and sand contained less organic carbon (460 and 180 times less, respectively) than deep-sea sediments prior to incubation (Table 2). After 1 year, in both artificial sediments (without and with chitin) the organic carbon content was significantly higher than at the start of the experiment.
| control (days) | chitin-enrichment (days) | ||||
|---|---|---|---|---|---|
| 0 | 7 | 365 | 7 | 365 | |
| deep-sea sediment | 0.935 ± 0.029 (A) | 0.989 ± 0.009 (A) | 1.670 ± 0.050 (A) | 1.889 ± 0.101 (A) | 2.143 ± 0.171 (A) |
| glass beads | 0.002 ± 0.002 (E) | 0.005 ± 0.003 (D) | 0.043 ± 0.006 (C) | 0.008 ± 0.002 (D) | 0.174 ± 0.041 (B) |
| sand | 0.005 ± 0.001 (D) | 0.007 ± 0.001 (D) | 0.058 ± 0.001 (C) | 0.009 ± 0.004 (D) | 0.065 ± 0.011 (C) |
- All data are given as means of three repetitions ± SD.
- Different letters (in parentheses) indicate significant differences between mean treatments as determined by post hoc Tukey’s HSD tests at P < 0.05.
Effects of sediment type, chitin enrichment and incubation time on bacterial community structure
Results from T-RFLP analysis in control and chitin-enriched natural and artificial sediments at different incubation times are summarised in Fig. 5. The total number of terminal restriction fragments (hereafter indicated as T-RFs) observed in all samples investigated was 148. Statistical analysis indicated that the numbers of T-RFs per sampling time were not significantly different as determined by post hoc Tukey’s HSD tests at P < 0.05. In control sediments without chitin, the initial total number of T-RFs ranged from 54 (control deep-sea sediment) to 21 (control glass beads). The total number of T-RFs in chitin-enriched deep-sea sediment after 7 days was lower (37) than in control deep-sea sediment (51), whereas the number of T-RFs in chitin-enriched artificial sediments was comparable to the respective controls. After 1 year, the total number of T-RFs in artificial sediments slightly increased, regardless of sediment treatment. Thirty-one T-RFs were present exclusively in control sediments without chitin addition, whereas 35 T-RFs were present only in chitin-enriched sediments.
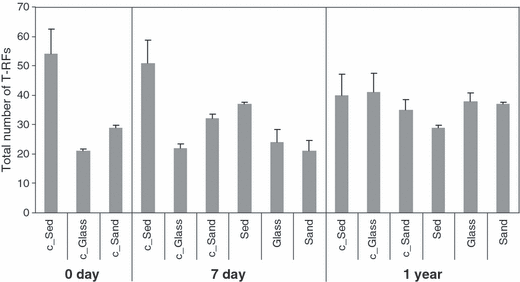
Total number of terminal restriction fragments (T-RFs) in control and chitin-enriched sediment types at different incubation time. The total number of T-RFs resulted from the separate digestion of amplified bacterial 16S rRNA genes with the endonucleases HhaI and MspI. Sediment (‘Sed’) indicates deep-sea sediments. Control sediments without chitin are labelled with the ‘c’ prefix. Bars indicate standard deviations.
Cluster analysis of T-RFLP data generally indicated that microbial community profiles from all sediment types collected after 1 year were more related to each other than those from shorter incubation times or from the control treatments (Fig. 6). Interestingly, within the no-chitin amended treatments, the more marked differences in community structure occurred after 1 year as well, generally regardless of the sediment type considered. In contrast, after 7 days’ incubation, the glass treatment with chitin seemed to cluster on its own, whereas the microbial structure of the natural sediment samples was more related to deep-sea sediment control treatments.
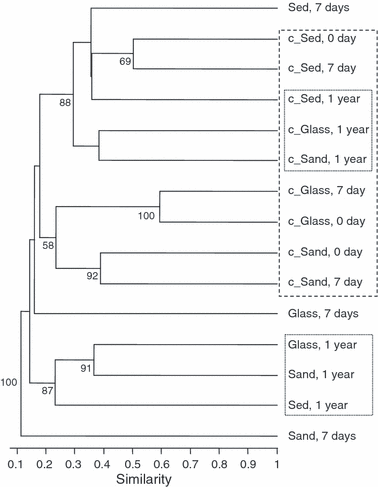
Cluster analysis of T-RFLP data of control and chitin-enriched sediment types at different incubation times. The UPGMA dendrogram was constructed based on T-RF presence/absence data obtained by T-RFLP analysis of the different sediment samples. Nodal support was assessed by 100 bootstrap replicates and only bootstrap values greater than 50% were shown. Sediment (‘Sed’) indicates deep-sea sediments. Control sediments without chitin are labelled with the ‘c’ prefix. See text for the description of clustering results.
These observations were corroborated by nMDS ordination of the T-RFLP data (Fig. 7), which, contrary to cluster analysis, does not assume a hierarchical structure among samples, but rather a gradual change (Ramette 2007). Sample points representing chitin-enriched sediments were found to be more spread in the nMDS plot with increasing incubation time. To assess the statistical significance between groups of samples based on their T-RFLP profiles we used the nMDS ordination in association with non-parametric ANOSIM tests. Control samples without additional chitin and samples amended with chitin fell into two overlapping (Fig. 7b) but significantly different groups (ANOSIM R = 0.326, P = 0.005). Short- as well as long-term incubation times, regardless of chitin addition or sediment type seemed also to induce community shifts with more pronounced differences after 1-year incubation (R = 0.33, P = 0.005; Fig. 7c). When samples were grouped by sediment type (natural sediments, glass beads and sand; Fig. 7d), the ANOSIM test also showed significant differences in bacterial assemblages (R = 0.247, P = 0.017). Because ANOSIM R values were generally below 0.5, those groups were most likely separated but overlapping to certain extent (Clarke & Gorley 2001), as may occur if the community structures changed over time but kept some common structure with their initial states (i.e. that at the beginning of the experiments).
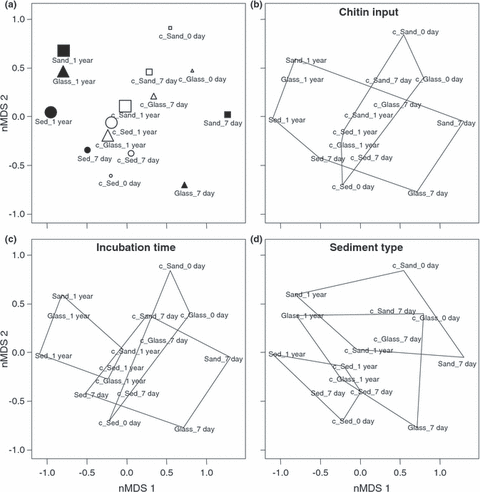
Non-metric multidimensional scaling (nMDS) ordination of T-RFLP data based on Jaccard dissimilarities among samples. (a) Symbols were assigned to each treatment as follows: squares, triangle and circle represent sand, glass beads, and natural sediment treatment, respectively, whereas full and empty symbols correspond to chitin-amended and not-amended controls (also with a ‘c’ prefix), respectively. The size of the symbols increases with incubation time of the respective samples. To further facilitate the interpretation of the nMDS results, geometric shapes were superimposed on the ordination. They depict (b) chitin-amended samples versus no-chitin control samples, (c) different incubation times (0, 7, 365 days), and (d) the three sediment types (deep-sea sediment, sand, glass beads). The overall stress of the ordination was 15.2%.
The variation in the bacterial community structure was further analysed by distance-based redundancy analysis (db-RDA) so as to determine which factors had the most significant effects on the variation of microbial assemblages. The respective effects of microbial cell number and biomass did not contribute significantly to the variation in T-RFLP data (P > 0.05; data not shown). The other three factors investigated (chitin input, incubation time and sediment type), however, each had significant effects on the variation in bacterial community patterns (P < 0.05; Fig. 4c). Similar to variation in chitobiase activity (Fig. 4a,b), sediment type was the most important factor and explained much more biological variation (13%) than did duration of incubation (8%) or chitin input (6%). The low amount of covariation among factors could indicate that the factors mostly behave independently from each other (Legendre & Legendre 1998). Although the three factors significantly explained the variation in the T-RFLP data, there was still a large amount of biological variation that could not be explained (i.e. 78%).
Discussion
There is a rich body of literature that has examined the general relationships between the nature of sediments and benthic microbiota. For instance, sediments composed of small particles with surface irregularities and high organic carbon content promote higher microbial attachment, and therefore higher colonisation and activity than sediments with coarser particles, having smooth surfaces and low organic carbon content (e.g.Hargrave 1972; Dale 1974; Nickels et al. 1981; DeFlaun & Mayer 1983; Meyer-Reil 1993; Mayer 1994). Those previous findings were mainly based on laboratory studies (e.g.Yamamoto & Lopez 1985;Findlay et al. 1992; Sharma & McInerney 1994;Köster et al. 2005), but colonisation patterns in laboratory microcosms may differ from field situations due to experimental artefacts (Targatz et al. 1983). The aim of our experiment was to investigate microbial colonisation (measured in growth and activity) due to different sediment types, ranging from fine to coarse sediments, after incubation directly at the deep-sea floor. Present results indicated that the structure and function of microbial deep-sea populations was clearly influenced by the nature of sediments (i.e. sediment particle size, particle shape, organic carbon content). We found significant relationships between decreasing particle size (from sand, glass beads, to deep-sea sediment, respectively), increasing organic carbon content and increasing microbial cell number, biomass and chitobiase activity. Regardless of chitin addition and incubation time, deep-sea sediments always exhibited higher values for microbial abundance, biomass and activity compared to the artificial sediments glass beads and sand. With the assumption that only a minor proportion of sediment organic carbon originates from bacterial carbon (0.6–8.5%; Dale 1974), we presume that, beside the gradual accumulation of sedimentary organic material, the enhanced bacterial production of extracellular chitinolytic enzymes in chitin treatments contributed to the formation of a complex organic matrix within sediments. This process of sediment stabilisation by microbial exudates could have also contributed to the further increase in organic carbon content. Despite this increase in organic carbon content in artificial sediments, values were always lower than those from natural sediments, supporting the finding that in fine-grained sediments the organic carbon content is generally higher than in sediments composed of larger particles (DeFlaun & Mayer 1983). It may thus also be hypothesised that the low carbon content limited microbial colonisation in the artificial sediments. Those observations are consistent with previous findings from laboratory experiments and were still observed after 1-year incubations under natural conditions.
Surface conditions of deep-sea sediments may frequently change due to disturbances such as near-bottom currents that can induce regular sediment transport or erode the sediment surface (Gage & Tyler 1991). The impact of benthic organisms and their activities can further affect the physical and chemical nature of the sediment surface including the abrasion, translocation and mixing of sediments (Findlay et al. 1990). The intensity of such processes and their impacts in deeper waters, however, may be enhanced if human disturbances such as commercial trawling (Kaiser 1998), mining (Radziejewska & Stoyanova 2000), and oil exploration (Jones et al. 2006) are superimposed. Maximal colonisation on such sediment surfaces will temporarily be reduced as a result of disturbances, until (re)colonisation by benthic organisms resumes. The present results may indicate that (re)colonisation of new sediments by deep-sea microbial communities after disturbance events accompanied by changes in physical or chemical nature of the sediment (e.g. sediment grain size or organic carbon content) apparently proceeds over a course of several months. This may also explain why a significant microbial response to the chitin supply was recorded only after the long-term experiment. Time lags of more than 1 week may be needed to observe enhanced enzyme production as a response to chitin pulses (Kanzog et al. 2009). Due to logistical limitations it was, however, not possible to obtain samples on a shorter timeframe. Our results at least identified some lower and upper time bounds within which significant changes in natural phenomena took place at the bottom of the Arctic Ocean.
The fact that sediment type was found to be the most important factor influencing chitobiase activity might be explained as follows. As a substantial fraction of the extracellular enzymes is bound to cell membranes or other surfaces (Meyer-Reil 1990; Martinez & Azam 1993), enzyme production could only be induced when microorganisms are present in the vicinity of nutrient particles (Karner & Herndle 1992). Thus, the degradation of chitin would be initiated only when cells come closer to the food source. For deep-sea sediments composed of finer particles it would thus be more likely that cells came into contact with the chitinous substrate. In addition, the diffusion of extracellular enzymes and monomers liberated by the enzymes away from the cells probably are more pronounced in coarse sediments than in sediments composed of smaller particles.
The structure of deep-sea microbial communities was also significantly influenced by all factors investigated. The analyses of T-RFLP profiles revealed that sediment type was apparently the most important factor explaining variation in community structure as compared to the effects of chitin input and incubation time. Although the three factors significantly explained the variation in the T-RFLP data, there was still a large amount of biological variation that could not be explained (i.e. 78%). Hence, additional environmental parameters would need to be further incorporated in the model to fully explain the changes in diversity patterns. Generally, results indicated that deep-sea sediments from the central HAUSGARTEN station harbour highly diverse bacterial assemblages. This is evident for the high number of different restriction fragments found in deep-sea sediment before the experiment (54 different bacterial ribotypes). Similar values of deep-sea benthic bacterial diversity, estimated using T-RFLP technique, have been also reported in previous studies (e.g.Urakawa et al. 2001; Luna et al. 2004; Quéric et al. 2008). The fact that the number of T-RFs in the artificial sediments glass beads and sand was higher after 1 year than in the initial samples may be explained by a gradual increase of the bacterial colonisation over the whole duration of the experiment. There are indications that, in contrast to the short-term incubation, bacterial communities had similar compositions after the long-term experiment for 1 year, regardless of sediment type. This interpretation should be made with caution as the present results were based on a limited dataset. Nevertheless, similar observations were reported by Findlay et al. (1992) during colonisation experiments with natural and artificial sediments in a controlled flow regime. The authors suggest that microbial communities developing in artificial barite sediment after a sufficient time would exhibit similar structure and biomass to communities present in ambient natural sediment.
Our results support the hypothesis that the shift in community structure with increasing incubation time is also influenced by the amendment of sediments with chitin. Virtually nothing is known about possible shifts in benthic deep-sea microbial community structure that could be associated with organic matter supply, although such shifts are evident in the community structure of megafauna and macrofauna organisms (e.g.Snelgrove et al. 1996; Billett et al. 2001). For example, in situ experiments using colonisation trays showed a fauna with high density but low diversity composed of more opportunistic species in organic enriched sediments, whereas control unenriched trays were found to host a low density but higher diversity fauna (Snelgrove et al. 1996). It is therefore reasonable to expect that chitin addition led to a decreased diversity and to a community which was dominated by few chitin-specific bacteria. Although this cannot be verified by the present data, a trend towards a reduction of T-RF richness only in deep-sea sediment enriched with chitin seems to corroborate this assumption. These observations would support the general hypothesis that increased food availability in an oligotrophic environment such as the deep sea may lead to a shift to more opportunistic species, increased dominance, and decreased diversity (Snelgrove et al. 1996).
Although our study was carried out in situ, under natural conditions at the sea floor, it is important to note that the artificial nature of tray experiments might still have modified natural conditions, e.g. by creating hydrodynamic biases (Smith 1985; Snelgrove et al. 1995). These tray-induced variations may also influence benthic biological processes such as sediment deposition, erosion and nutrient flow. Our sediment trays were approximately 20 cm above the sea floor, and therefore, sediments within the chambers inside the trays were disconnected from the surrounding sea floor. Hence, (re)colonisation mainly occurred by the already existing communities present in the samples and (re)suspended microorganisms transported by near-bottom currents. Another factor that we could not control for was grazing, which may have prevented microbial communities from reaching their full potential size after 1-year incubation. Indeed, although grids (5 mm mesh size) were applied to avoid the interference of larger animals, we could not exclude meiofauna (e.g. nematodes) or microeukaryotic organisms as potential grazers. Hence, microbial abundance and biomass could actually be higher than reported here. In addition, grazing may also have an effect on community structure, as with increasing sediment particle smoothness the susceptibility of bacteria to predation increases (Nickels et al. 1981). Although it is difficult to control for all experimental biases when working under in situ conditions, we assume that all treatments inside the chambers were similarly affected by those environmental conditions and that sediment trays allowed at least valid inter-treatment comparisons to be made. This approach may thus contribute to a better understanding of microbial colonisation processes under deep-sea conditions.
In conclusion, deep-sea microbial communities respond to variation in sediment types and chitin input by changing their composition, abundance and activity. In general, our observations support the idea that microbial communities in natural environments prefer to colonise sediments with smaller grain sizes and higher organic carbon contents. The effect of chitin was only seen after 1 year of incubation, which gives an upper time bound to observe significant changes in microbial response due to chitin input. Future studies will incorporate this knowledge and will address the identification of the microbes associated with high chitin degradation rates and of environmental variables that may contribute to the variation of microbial community structure in the Arctic Ocean.
Acknowledgements
We are grateful to the ship officers and crew of the German research-icebreaker Polarstern for their help during the expeditions to HAUSGARTEN in summer 2004 and 2005. Experiments with the free-falling colonisation frame would not have been possible without the support of J. Wegner and I. Schewe. We are also indebted to I. Kolar and M. Volkenandt, for assistance with the sample preparation, biochemical analyses and bacterial counts. We wish to thank A. Boetius, T. Soltwedel, and three anonymous reviewers for useful comments and discussions.




