In situ localization of the evolutionary conserved Cpy/Cty gene in the subfamily Chironominae (Chironomidae, Diptera): establishment of chromosomal homologies
Abstract
The homologous sites on the salivary gland chromosomes of 13 species from three genera: Chironomus, Glyptotendipes, Kiefferulus have been mapped by means of fluorescent in situ hybridization using the evolutionary conserved gene Cpy/Cty (clone Cla1.1). In all species of genus Chironomus and genus Kiefferulus, the Cty/Cpy gene is located on arm F of chromosome EF. The relocation of the gene among the species of genus Chironomus can be done by simple or complex homozygous inversions which occurred during the divergent evolution of the chromosome of the species. In the genus Glyptotendipes, the Cty/Cpy gene was localized in arm E of chromosome EF. Since the banding patterns of salivary gland chromosomes between genus Chironomus and genus Glyptotendipes cannot be compared directly, in situ hybridization with clone of conservative gene was performed to be established some homologous chromosomes. The results obtained indicate that the chromosome arm F of Chironomus and chromosome arm E of Glyptotendipes may be homologous.
Introduction
For tracing the chromosome rearrangements involved in the evolution process several approaches were proposed (Bondinas et al. 2001): specifically by examining the banding patterns of larval polytene chromosomes and analyzing the configuration of hybrid polytene chromosomes. The latter allows the identification of both chromosome homologies and rearrangements by which the species differ (Michailova 1989a). In genus Drosophila other approach was applied –in situ hybridization of labelled DNA sequences to their complementary DNAs of the polytene chromosomes of different species (Steinemann et al. 1984; Loukas and Kafatos 1986; Bondinas et al. 2001). This is a suitable method for the identification of homologous regions of the chromosomes providing additional evidence to those obtained by classical approaches. Up to now this approach has not yet been applied to chironomids. In the present study, in situ hybridization using the evolutionary conserved gene Cpy/Cty was performed. We used clone Cla1.1, which contains the gene Cty that is closely linked to the male-specific Cla element cluster hybridized unequivocally and preferentially within the sex determining region of Chironomus thummi Meigen, 1804 (Kraemer and Schmidt 1993). We examined the localization of Cpy/Cty gene in seven species of the Chironomus, five species of Glyptotendipes and one species of Kiefferulus. The aim was to identify homologous loci on the polytene chromosomes of different chironomid species. Since the Cty/Cpy gene is a single copy gene, one can be rather sure that the Cpy/Cty locus is homologous in all chironomids. The location of this gene in different cytocomplexes of Chironomus, sibling species of Glyptotendipes as well as in a species of Kiefferulus can be taken as a reliable tag for the identification of homologous chromosomal regions. Since the Cty/Cpy gene is extremely well conserved among species (Kraemer et al. 2000), we decided to use this gene to map homologous regions even in different genera.
Materials and Methods
Taxa considered in this study
We used seven species of the genus Chironomus belonging to two cytocomplexes: thummi and pseudotummi, five species of Glyptotendipes and one species of Kiefferulus (Table 1). All but one species of Chironomus and Kiefferulus were collected from the nature, while with the exception of Glyptotendipes barbipes, all other species of Glyptotendipes were from our laboratory cultures.
| Genus | Species | Localities | Time of collecting |
|---|---|---|---|
| Glyptotendipes | G. salinus | Lab. culture | 06.1999 |
| G. barbipes | Germany | 1986 | |
| G. pallens | Lab.culture | 07.1999 | |
| G. glaucus | Lab. culture | 07.1999 | |
| Glyptotendipes sp. (a hybrid, Michailova, 1998) | Lab. culture | 1998 | |
| Chironomus | C. plumosus | Pazardzik (Tri Vodici) | 20.06.2002 |
| C. balatonicus | Bourgs Lake | 23.02.2002 | |
| C. nuditarsis | Lom (Rasovo) | 16.05.2002 | |
| C. luridus | Sofia | 09.09.2001 | |
| Chironomus sp. | Lab. culture | 09.09.2002 | |
| C. annularius | Sofia (Bezden) | 04.06.2002 | |
| C. salinarius | Atanasovso Lake | 10.10.2003 | |
| Kiefferulus | K. tendipediformis (Cytotype 2) | Olkusz (near Krakow), Poland | 06.2002 |
In situ hybridization
The biotin as well as the digoxigenin labelled clone DNA was localized in the polytene chromosomes by in situ hybridization as described for Chironomus by Schmidt et al. (1988). After hybridization and detection of the signals, chromosomal preparations were stained with aceto–orcein and photographed once again in order to map precisely the position of the signal to the standard chromosomal banding patterns.
The chromosomes were identified and the gene was localized to a numbered division using the photographic maps of Keyl (1962), Devai et al. (1983), Kiknadze et al. (1991) for genus Chironomus and the maps of Walter (1973), Michailova (1989a,b), Michailova (1998), Michailova and Contreras-Lichtenberg (1995) for genus Glyptotendipes. Kiefferulus tendipediformis chromosomes were identified and the hybridization signal was established using the photographic map of Michailova et al. (2005).
Results
Glyptotendipes
In situ hybridization of the clone Cla1.1 containing the Cty gene to the polytene chromosomes of all species of Glyptotendipes showed a distinct signal over one chromosomal band on chromosome EF, arm E, near the telomere region. The signal is localized in different section of this region (Fig 1a,b) (Table 2).
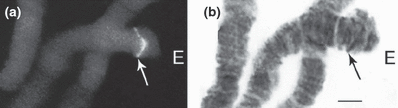
Fluorescence and bright field micrographs of chromosome arm E of Glyptotendipes salinus. (a) Strong fluorescence after in situ hybridization with the conserved Cpy/Cty gene at the site corresponding to region 1a, according to the G. salinus map of Michailova (1989b). (b) Bright field micrograph of the same chromosome arm (aceto–orcein staining). Bar represents 10 μm
| Species | Cytocomplex and cytotype | Arm | Numbering of the arm | According to the authors | Figures |
|---|---|---|---|---|---|
| G. salinus | – | E | 1a 2 3 4 5 | Michailova (1989b) | Fig. 1a,b |
| G. barbipes | – | E | 1a 2 3 4 5 | Michailova (1989b) | Not shown |
| G. pallens | – | E | 1 2a 3 4 5 | Michailova and Todorova (1998) | Not shown |
| G. glaucus | – | E | 1 2a 3 4 5 | Michailova and Contreras-Lichtenberg (1995) | Not shown |
| Glyptotendipes sp. | – | E | 1 2a 3 4 5 | Michailova (1998) | Not shown |
| C. balatonicus | thummi | F | 1 2 3 4 5 6 7 8 9 10 17 16 15 14 13 12 11 18 19 20 21 22 23 | Devai et al. (1983) | Fig. 2a,b |
| Chironomus sp. | pseudothummi | F | 1 2 3 4 5 6 7 8 9 10 15 14 13a-c-d 12 11 16 17 18-23 | Keyl (1962) | Not shown |
| C. annularius | thummi | F | 1 2 3 4ab 8c-a 7 6 5 4dc 17 16 15 14 13 12 11 10 9f-c 8ed 18-23 | Keyl (1962) | Not shown |
| C. nuditarsis | thummi | F | 1a-d 6 5 4 3 2 1 8c-a 7 1e-g 8d-f 9 10 17 16 15 14 13 12 11 18-23 | Keyl (1962) | Fig. 3a,b |
| C. plumosus | thummi | F | 1a-d 6 5 4 3 2 1 7 8 9 10 17 16 15 14 13 12 11 18-23 | Keyl (1962) | Not shown |
| C. luridus | pseudothummi | F | 1a-h 9 8 7 6 5 4 3 2 1 13c-a 12 11 16a-e 10 15 14 13d 16f-g 17 18-23 | Keyl (1962) | Not shown |
| C. salinarius | Unclear position | F | Not far from the centromere | Keyl and Keyl (1959) | Fig. 4a,b |
| K. tendipediformis | Cytotype 2 | F | Middle of arm F | Michailova et al. (2005) | Fig. 5a,b |
- Bold indicates the location of the signal.
Chironomus
In both Chironomus cytocomplexes studied (thummi and pseudothummi) the hybridization signal is localized in chromosome arm F, section 1 (2, 3) (Table 2). In the studied species, position of the section 1 is changed due to fixed homozygous inversions. In Chironomus salinarius the signal is close to the centromere region (Fig. 4a,b).
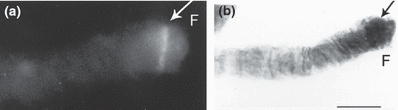
Fluorescence and bright field micrographs of chromosome arm F of Chironomus balatonicus. (a) Strong fluorescence after in situ hybridization with the conserved Cpy/Cty gene at the site corresponding to region 1, according to the C. balatonicus map of Devai et al. (1983). (b) Bright field micrograph of the same chromosome arm (aceto–orcein staining). Bar represents 10 μm
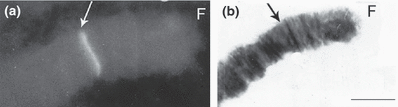
Fluorescence and bright field micrographs of chromosome arm F of Chironomus nuditarsis. (a) Strong fluorescence after in situ hybridization with the conserved Cpy/Cty gene at the site corresponding to region 1, according to the C. nuditarsis map of Keyl (1962). (b) Bright field micrograph of the same chromosome arm (aceto–orcein staining). Bar represents 10 μm
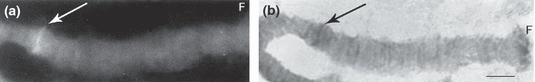
Fluorescence and bright field micrographs of chromosome arm F of Chironomus salinarius. (a) Strong fluorescence after in situ hybridization with the conserved Cpy/Cty gene at the site not far from the centromere. (b) Bright field micrograph of the same chromosome arm (aceto–orcein staining). Bar represents 10 μm
Kiefferulus
Kiefferulus tendipediformis belongs to cytotype 2 (2n = 6) (Michailova et al. 2005). The Cpy/Cty gene hybridizes in the middle of arm F close to the centromere region (Fig. 5a,b).
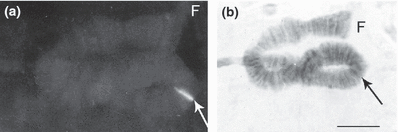
Fluorescence and bright field micrographs of chromosome arm F of Kiefferulus tendipediformis. (a) Strong fluorescence after in situ hybridization with the conserved Cpy/Cty gene at the site in the middle of the arm. (b) Bright field micrograph of the same chromosome arm (aceto–orcein staining). Bar represents 10 μm
Discussion
The gene Cpy/Cty shows conservation of its position in the chromosomes of the studied genera. In all studied Chironomus species and in K. tendipediformis, the Cpy/Cty gene is located in chromosome arm F. The detailed cytogenetical analysis in genus Chironomus showed that intrachromosomal rearrangements (simple and complex homozygous inversions) have moved the Cpy/Cty gene from the telomere in Chironomus balatonicus and Chironomus sp. to proximal position in Chironomus plumosus, Chironomus nuditarsis, Chironomus luridus. In K. tendipediformis the locus is found further along arm F than in the above species, while in C. salinarius the locus occur closer to the centromere of the chromosome. The banding patterns of these species were not sufficiently distinct for precise identification of homologous banding patterns in comparison with the other species of Chironomus. These differences might be due to a larger number of chromosome rearrangements during the divergent evolution of these two species.
In Glyptotendipes the Cpy/Cty gene is found on arm E. It should be noted that we used in our study for Glyptotendipes the cytological nomenclature of chromosome arms of Walter (1973), which is different from the one used for the genus Chironomus in which chromosomal homologies are well established (Keyl 1962). The differences in banding patterns between the genus Glyptotendipes and Chironomus are so extensive that only few, short sections may be homologized. Therefore it is problematic to establish unequivocally cytogenetic relationship between species belonging to these different genera. On other hand, it is widely accepted that chromosomal loci hybridizing with the same single copy DNA probe are homologous with high probability (Loukas and Kafatos 1986). Taking the result of the hybridization of the conserved Cpy/Cty gene into consideration, then it is improbable that the chromosome arm F of genus Chironomus and chromosome arm E of genus Glyptotendipes could be homologous.
Acknowledgement
This work was supported by grant No. B1601 from Ministry of Education and Science, Sofia, Bulgaria.




