Signal transducer and activator of transcription 1 negatively regulates constitutive gamma interferon-inducible lysosomal thiol reductase expression
Senior author: Maja Maric
Summary
Gamma interferon-inducible lysosomal thiol reductase (GILT) is an enzyme that catalyzes thiol bond reduction and plays an important role in the early steps of antigen processing. The key factor involved in the regulation of GILT expression upon cell stimulation with interferon-γ (IFN-γ) is signal transducer and activator of transcription 1 (STAT1). In this study, we examined the role of STAT1 in regulating the constitutive expression of GILT. We showed that STAT1 interacts with the GILT promoter, even in the absence of IFN-γ, and that STAT1 represses GILT expression. These results reveal an atypical negative regulatory role for STAT1 in the constitutive regulation of genes involved in antigen processing.
Introduction
Thiol reductases are enzymes that carry out the oxido-reduction of disulphide bonds in proteins.1 They are located in various cellular compartments such as the mitochondria,2 endoplasmic reticulum3 and lysosomes.4–6 Gamma interferon-inducible lysosomal thiol reductase (GILT) is a unique thiol reductase that reduces disulphide bonds under the low-pH conditions found within lysosomes. Through the reduction of thiol bonds of endocytosed proteins, GILT unfolds native protein antigens in preparation for subsequent processing by lysosomal proteases.
The mature form of GILT is a 30 000 molecular weight (MW) enzyme, which has the conserved active-site motif (-CGAC-)1,3 typical of thiol reductases. However, it functions at an acidic pH of 4·5–5·5, and thus differs from other thiol reductases that function at neutral or alkaline pH.7 GILT is constitutively expressed in professional antigen-presenting cells (APCs),8 but also in T cells9 and fibroblasts.10 GILT is also secreted in tissue culture supernatants of GILT-expressing B-cell lines.11 GILT protein expression is moderately increased upon treatment with interferon-γ (IFN-γ). It has been shown that upon IFN-γ stimulation the expression of GILT is regulated by signal transducer and activator of transcription 1 (STAT1) in human melanoma cells.12Stat1 is one of the seven members of a family of STATs – latent cytoplasmic proteins activated by various stimuli (cytokines and growth factors) and involved in the regulation of cell growth and differentiation, immune response and homeostasis.13
Stimulation with IFN-γ results in the activation of Janus kinases (Jak) 1 and 2. Activated Jaks phosphorylate tyrosine residues on the IFN-γ receptor, which serve as STAT1 docking sites. Following phosphorylation of tyrosine 701 (Y701) STAT1 monomers homodimerize, translocate to the nucleus and activate the transcription of target genes14–16 through binding to γ-activated sequence elements (GAS).17 The promoters of IFN-γ-activated genes usually contain GAS.13 Two putative GAS sequences have been identified in the GILT promoter at 130 and 510 bp upstream of exon 1 of the GILT gene.
There are two naturally occurring forms of STAT1: STAT1α and the alternatively spliced isoform STAT1β. STAT1β lacks the 38 amino acid residues in the C-terminal transcriptional activation domain that can bind the histone acetyltransferases p300/CBP.18,19 STAT1 is primarily activated through phosphorylation at tyrosine 701.20 A secondary, independent, phosphorylation event occurs at serine 727, which is needed for maximal transcriptional activity.21
In addition to its role in regulating the expression of target genes upon stimulation with IFN, STAT1 has also been shown to play a role in the constitutive expression of certain genes: low Molecular mass Polypeptide 2 (LMP2),22,23 caspases24 and major histocompatibility complex (MHC) class I.25
In this study, we investigated whether STAT1 interacts with the GILT promoter in the absence of IFN-γ. Our data suggest that the presence of Stat1 in a mouse fibroblast cell line correlates with decreased activity of the GILT promoter and decreased constitutive expression of GILT protein. The DNA affinity precipitation assay (DAPA) showed that STAT1 binds with high specificity to putative GAS motifs in the GILT promoter in the absence of IFN-γ stimulation. We also showed that STAT1 residues Y701 and S727 are not required for constitutive STAT1 binding to the GILT promoter. Therefore, phosphorylation of Y701, thought to be necessary for STAT1 homodimerization, is not required for constitutive binding of STAT1 to the GILT promoter. The absence of C-terminal amino acids from the alternatively spliced form of STAT1β does not prevent the binding of STAT1 to the GILT promoter. The remaining N-terminal portion of STAT1 seems to be crucial for binding of STAT1 to the GILT promoter, independently of IFN-γ stimulation. Our experiments indicate that STAT1 residues 426/427 are required for constitutive interaction of STAT1 with the GILT promoter.
Materials and methods
Cell lines and plasmids
Wild-type (WT) and Stat1−/− mouse embryonic fibroblasts (MEFs) were obtained from Dr David Levy (New York University School of Medicine, New York, NY). Cells were maintained in Dulbecco’s modified Eagle’s minimal essential medium (Invitrogen, Frederick, MD) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT).
Stat1 constructs (Stat1α and Stat1β) were a kind gift from Dr D. Levy, New York University Medical Center, NY. Stat1α-Y701F, Stat1α-S727A, Stat1α-Y701F/S727A and Stat1β-Y701F were generated by site-directed mutagenesis using the QuikChange mutagenesis kit (Agilent, Santa Clara, CA). Constructs were subcloned into the pcDNA 3.1+ plasmid which carries the hygromycin resistance gene (Invitrogen). Transfections were carried out using Lipofectamine LTX (Invitrogen) according to the manufacturer’s protocols. Stable transfectants were selected and maintained in medium supplemented with 400 μg/ml of hygromycin (Invitrogen). All constructs were verified by sequencing (Genewiz, South Plainfield, NJ).
IFN--γ stimulation, Western blot analysis and antibodies
Cells were stimulated with mouse IFN-γ (100 μ/ml; Peprotech, Rocky Hill, NJ) for 24 hr and whole-cell protein extracts were prepared with the addition of protease inhibitors (Roche Diagnostics, Nutley, NJ) and phosphatase inhibitor cocktails 1 and 2 (Sigma-Aldrich, St. Louis, MO). Protein quantification was carried out using the bicinchoninic acid (BCA) assay (Pierce, Rockford, IL). For Western blotting to detect GILT protein, 5 μg/lane of protein extract was loaded onto 15% sodium dodecyl sulphate (SDS)-polyacrylamide gels. Proteins were transferred onto poly(vinylidene difluoride) (PVDF) membranes. Primary antibodies used for detection were GILT (rabbit polyclonal antiserum; M. Maric), actin (Sigma-Aldrich), total STAT1 (Cell Signaling, Danvers, MA). Anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody (Jackson Immunoresearch, West Grove, PA) was used. Detection was carried out using the ECL plus reagent (PerkinElmer, Gwaitham, MA).
Biotin-labelled DNA affinity precipitation assay
The sequences of the 5′ biotinylated oligonucleotides (IDT, San Diego, CA) used for the DNA affinity precipitation assay (DAPA) were as follows: STAT1 GAS Site Probe 1, GCGGAGCCTTCAGGAAAGGAGTCCCAGG and STAT1 GAS Site Probe 2, CACACTCAGTTGCTGGAAGCAAGTACCTCA; and the non-biotinylated oligonucleotides used were Stat1 consensus, TCGAGCCTGATTTCC-CCGAAATGAGGC and p53, TCCGAACAAGTCCGGGCATATGT.
Complementary oligonucleotides were mixed with the above-mentioned sequences and annealed. Five-hundred micrograms of whole-cell lysate was incubated with 900 pmol of biotinylated oligonucleotide, and the complex was immunoprecipitated using streptavidin-conjugated agarose beads (Millipore, Temecula, CA), based on a previously described protocol.12 Oligonucleotide competition assays were performed using either a 10-fold or a 50-fold excess of nonbiotinylated DNA oligonucleotides. Proteins were eluted from streptavidin-conjugated agarose beads and analyzed by Western blotting, after SDS-PAGE (12% gel).
Luciferase assay
A 772-bp fragment of the GILT promoter was cloned into the pGL3 Enhancer and pGL3 Basic plasmids (Promega, Madison, WI), fused to the firefly luciferase reporter. Empty vectors were used as controls. The plasmids were transfected into WT and Stat1−/− cells using Lipofectamine LTX (Invitrogen). In some cases, luciferase plasmids were co-transfected with various Stat1 constructs, into Stat1−/− cells. pRL-SV40 (Promega) encoding Renilla luciferase, was co-transfected at a luciferase : firefly ratio of 1:10. Whole-cell lysates were prepared 48 hr post-transfection, and the assay was carried out using the dual-reporter luciferase assay kit (Promega). Samples were read on a Berthold luminometer. Luciferase values were normalized to Renilla expression for each sample.
Results
Increased expression of GILT protein in Stat1−/− fibroblasts
Typically, STAT1 regulates gene expression upon stimulation with IFN, but STAT1 has been also implicated in regulating the constitutive expression of several genes.22–25 Thus, we tested whether STAT1 would have an effect on the constitutive expression of GILT. We hypothesized that the lack of STAT1 regulation in Stat1−/− MEFs would either not affect the constitutive expression of GILT or would decrease it when compared with WT MEFs.22,24Stat1−/− MEFs19,26 and WT MEFs were tested for the expression of GILT by Western blotting. Surprisingly, semiquantitative Western blot analysis of Stat1−/− MEFs showed an increased expression of GILT protein that was not dependent on IFN-γ treatment (Fig. 1a). When WT MEFs were treated with IFN-γ, GILT expression was increased (Fig. 1b), whereas the levels of GILT in IFN-γ-treated Stat1−/− MEFs remained unchanged. These MEFs were derived from C57BL/6 mice. The same result was achieved using MEFs derived from CD1 mice (data not shown), therefore excluding the possibility that this phenotype is specific to this particular fibroblast cell line.
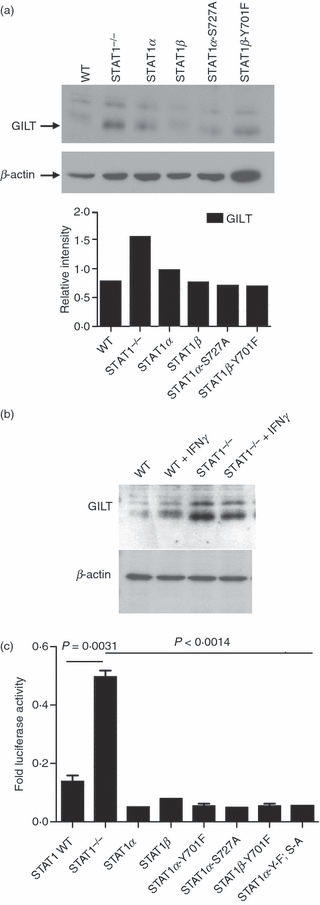
(a) Levels of gamma interferon-inducible lysosomal thiol reductase (GILT) are up-regulated in signal transducer and activator of transcription 1 (Stat1)−/− mouse embryonic fibroblasts (MEFs). Whole-cell lysates were prepared from wild-type (WT) Stat1 cells, Stat1−/− cells and Stat1−/− cells stably transfected with Stat1α, Stat1β, Stat1α-S727A and Stat1β-Y701F. The protein concentrations were determined by the bicinchoninic acid (BCA) assay. Five micrograms of whole-cell lysate was loaded into each lane of 15% sodium dodecyl sulphate (SDS)-polyacrylamide gels. After electrophoresis, the separated proteins were transferred to a poly(vinylidene difluoride) (PVDF) membrane. The blots were probed with anti-GILT, and the bound antibody was revealed using chemiluminescence. The membranes were then stripped and reprobed with β-actin-specific antibody as a loading control. (b) Treatment with interferon-γ (IFN-γ) increases GILT expression in WT, but not in Stat1−/− MEFs. WT and Stat1−/− MEFs were treated for 24 hr with 100 U/ml of IFN-γ. Cells were lysed in Tris saline buffer containing TX-100 and protease inhibitors. Equal amounts of total protein were separated by electrophoresis on a 12% SDS-polyacrylamide gel and transferred onto nylon membrane. The membrane was incubated with anti-GILT serum and horseradish peroxidase (HRP)-conjugated anti-IgG. The membrane was stripped and incubated with anti-actin as a loading control. Antibody binding was detected by chemifluorescence. (c) STAT1 decreases GILT promoter activity. WT and Stat1−/− cells were transfected with pGL3 basic+GILT and pRL SV40 at a 1:10 ratio. Stat1−/− cells were also transiently transfected with Stat1α, Stat1β, Stat1α-Y701F, Stat1α-S727A, Stat1β-Y701F and Stat1α-YF/SA, along with pGL3 basic+GILT and pRL SV40. The cells were harvested 48 hr post-transfection and assayed for luciferase activity. The results are expressed as Firefly luciferase activity normalized to Renilla luciferase activity, and are representative of three independent experiments performed in duplicate. The unpaired, two-tailed t-test was used to determine the P-value. A P of < 0·05 indicates a significant difference between the means.
Activation of GILT promoter in Stat1−/− cells
Increased expression of GILT protein in Stat1−/− MEFs led to the hypothesis that STAT1 may actually play a negative role in regulating the GILT promoter activity under basal conditions. To address this possibility, we used the luciferase assay to determine the specific activation of the GILT promoter in WT and Stat1−/− MEFs. The GILT promoter, 772 bp in length, was cloned into the pGL3 basic vector encoding the firefly luciferase reporter gene. The activity of the firefly luciferase reporter gene under control of the GILT promoter in WT cells and in Stat1−/− cells is shown in Fig. 1c. The decreased expression of GILT in unstimulated WT MEFs implies that phosphorylation of STAT1 is not required for the negative regulatory function of STAT1. Therefore, we transfected Stat1−/− cells with alternatively spliced forms of Stat1 (Stat1α and Stat1β), as well as with the phosphorylation-deficient mutants Stat1α-Y701F, Stat1α-S727A and Stat1β -Y701F, and the double mutant Stat1α-YF/SA, along with firefly luciferase plasmids expressing the GILT promoter. Our data show that the expression of any variant of Stat1 in Stat1−/− MEFs reduced the firefly luciferase activity controlled by the GILT promoter to levels similar to those seen in WT MEFs (Fig. 1c). These results suggest that the C-terminal transactivation domain and the phosphotyrosine-mediated dimerization, are not important for the regulation of constitutive GILT expression.
The remaining portion of STAT1 includes the DNA-binding domain,27,28 which may be responsible for constitutive binding of STAT1 to the GILT promoter. Previously, several groups have shown that the mutation of specific amino acids within the DNA-binding and linker regions in Stat1 can affect Stat1 binding and nuclear retention.29–31 Thus, we generated three Stat1 constructs mutated at DNA-binding sites and tested them in the luciferase reporter gene assay. The first mutant, Stat1-V426D/T427D, is defective in IFN-γ-induced Stat1 DNA binding to specific GAS sites and also shows weakened, non-specific protein–DNA interactions.29 The DNA-binding-deficient Stat1 mutant, E428A/E429S, has been shown to be tyrosine phosphorylated in response to IFN-γ and can be translocated to the nucleus, but cannot induce activation of the reporter gene.30 The third DNA-binding mutant, Stat1-K544A/E545A, previously characterized by Darnell et al.,31 has been shown to have increased off-rates from GAS sites. Hence, this mutant is present at the GAS sites for much shorter times than the WT protein but has been found to accumulate within the nucleus upon IFN-γ stimulation.29Stat1−/− and WT MEFs were co-transfected with a firefly luciferase reporter gene under the control of GILT promoter and either WT Stat1α or one of the three described DNA-binding mutants. Expression of either Stat1α (Fig. 2a) or two of the DNA-binding mutants (E428A/E429S and K544A/E545A) (data not shown) in Stat1−/− cells, decreased the luciferase activity. However, the cells transfected with the DNA-binding mutant V426D/T427D behaved like Stat1−/− cells, suggesting that this particular site is important for constitutive binding of STAT1 to GILT promoter in MEFs.
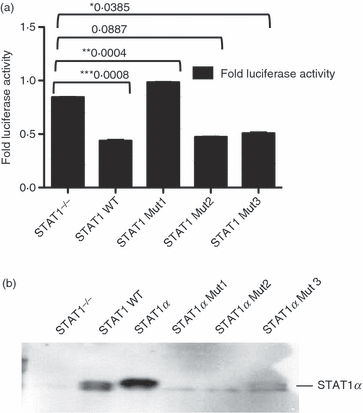
(a) DNA-binding mutants E428A/E429S and K544A/E545A decrease the gamma interferon-inducible lysosomal thiol reductase (GILT) promoter activity, but mutant V426D/T427D does not. Wild-type (WT) and signal transducer and activator of transcription 1 (Stat1)−/− cells were transfected with pGL3 basic+GILT and pRL SV40 at a 1:10 ratio. Stat1−/− cells were also transiently transfected with Stat1α and a DNA-binding mutant, Stat1-V426D/T427D. The luciferase assay was carried out as described in the text. Stat1α Mut1, V426D/T427D; Stat1α Mut2, E428A/E429S; and Stat1α Mut3, K544A/E545A. The unpaired, two-tailed t-test was used to determine the P-value. A P of < 0·05 indicates a significant difference between means, as denoted by asterisks * to ***. (b) The mutant V426D/T427D does not bind the GILT promoter in vitro. Lysates from WT, Stat1−/− and three mutant Stat1−/− mouse embryonic fibroblasts (MEFs) were incubated with biotinylated Stat1 oligonucleotides corresponding to the putative γ-activated sequence element (GAS) #2 present in the GILT promoter. The same experiment was carried out for putative GAS #1, and the same outcome was observed (data not shown). Proteins were immunoprecipitated using streptavidin–agarose beads, electrophoresed on SDS-polyacrylamide gels and analyzed by Western blotting. Binding of STAT1 to GAS sites was detected in WT cells, Stat1−/− cells transfected with Stat1α, and Mutant 3 but not in Stat1−/− cells transfected with Mutants 1 and 2.
In vitro interaction of STAT1 with the GILT promoter
Promoter regions of IFN-γ-inducible genes usually have a conserved nucleotide sequence, TTNCNNAA, known as the GAS, which directs rapid transcriptional activation upon Stat1 binding.28 Therefore, the mouse GILT promoter was analyzed for transcription of GAS sites using the Matinspector program.32 Two putative GAS sites were identified (Fig. 3a). Biotinylated oligonucleotides corresponding to these two sequences – STAT1 GAS Site Probe 1 (GCGGAGCCTTCAGGAAAGGAGTCCCAGG) and STAT1 GAS Site Probe 2 (CACACTCAGTTGCTGGAAGCAAGTACCTCA) – were tested for their ability to bind Stat1 in DAPA.33 These oligonucleotides were incubated with whole-cell lysates from WT or Stat1−/− MEFs (Fig. 3b). In order to confirm the specificity of binding, lysates from Stat1−/− and WT MEFs were also tested for binding in the presence of excess non-biotinylated competitors: either with excess Stat1 consensus sequence or with excess of a non-specific p53 oligonucleotide (Fig. 3c).
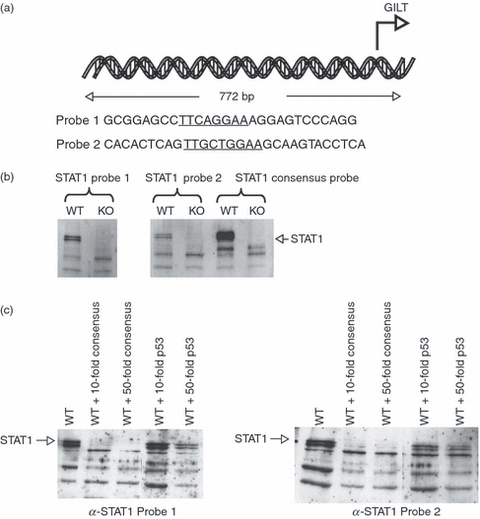
Signal transducer and activator of transcription (STAT1) binds to γ-activated sequence element (GAS) sites present in the gamma interferon-inducible lysosomal thiol reductase (GILT) promoter. (a) Putative STAT1-binding sites in the GILT promoter. GAS #1 is located 130 bp upstream of the first transcribed exon of GILT and GAS #2 is located 510 bp upstream. (b) STAT1 binds to putative GAS sites in the GILT promoter. Lysates from wild-type (WT) and Stat1−/− mouse embryonic fibroblasts (MEFs) were incubated with biotinylated Stat1 sequence oligonucleotides corresponding to the GAS sequences present in the GILT promoter. Proteins were immunoprecipitated using streptavidin–agarose beads, electrophoresed on SDS-polyacrylamide gels and analyzed by Western blotting. Binding to GAS sites was detected in WT cells, but not in Stat1−/− knockout (KO) cells, indicating the interaction of Stat1 with the GILT promoter. (c) STAT1 binding to the GILT promoter is specific. The experiment was performed as described in the text above. The specificity of the STAT1 interaction with the GAS site was demonstrated by the addition of excess unlabelled Stat1 consensus sequence or sequence from a p53-binding site. The latter lacks a GAS motif and cannot outcompete STAT1 binding to the GILT promoter.
Binding of STAT1 to biotinylated GAS site oligonucleotides corresponding to the sequence in the GILT promoter was successfully competed-out with only a 10- or 50-fold excess of Stat1 consensus sequence. Excess p53-binding nucleotide, which does not contain a GAS sequence, did not compete-out the binding of STAT1. Therefore, our data suggest that constitutive STAT1 binding to the GILT promoter occurs at GAS sites.
In addition, we tested whether mutations that affect the activity of the GILT promoter can influence in vitro binding to the GAS sites in the GILT promoter. The results shown in Fig. 2b indicate that mutant K544A/E545A (Mut 3) binds to the GILT promoter but mutant V426D/T427D (Mut 1) does not bind GAS sequences in GILT promoter, as expected. However, repeated DAPA did not detect binding of E428A/E429 (Mut 2), although this mutant behaved like STAT1α in the luciferase assay. This may be a result of either the limit of detection of DAPA or because this mutant exerts its effect on the GILT promoter indirectly.
Unphosphorylated STAT1 binds with specificity to the GILT promoter
To determine whether mutant STAT1 interacts with the specific sequences in the GILT promoter, regardless of the phosphorylation, WT, Stat1−/−, Stat1β-Y701 and Stat1α-S727 MEFs were treated with IFN-γ and the lysates were incubated with biotinylated oligonucleotides of Stat1 Probe 1 and Probe 2 (Fig. 4a). Our data indicate that, regardless of phosphorylation of Y701 and S727, STAT1 is able to bind target sequences in the GILT promoter. However, to confirm that what is seen here is specific binding, lysates from Stat1−/− cells transfected transiently with Stat1α, Stat1β-Y701 and Stat1α-S727 were incubated with biotinylated oligonucleotides of Stat1 Probe 1 and Probe 2 (Fig. 4b). The reactions were competed-out with a 50-fold excess of unlabelled probe corresponding to either Stat1 consensus or p53 sequences. Our data indicate that WT and Stat1 mutants can bind specifically to the sequences in the GILT promoter. Similar results were achieved with the Stat1 probe 1 (data not shown).
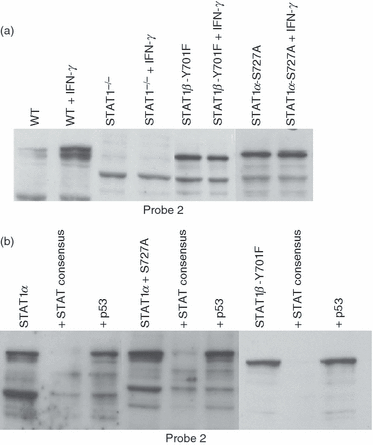
Binding of signal transducer and activator of transcription 1 (STAT1) to the gamma interferon-inducible lysosomal thiol reductase (GILT) promoter occurs independently of phosphorylation and is specific. (a) STAT1 binds to the GILT promoter in the presence or absence of interferon-γ (IFN-γ). Lysates from wild-type (WT) and Stat1−/− and from the stable transfectants Stat1α, Stat1α S727A and Stat1β-Y701F, stimulated with 100 U/ml of IFN-γ for 1 hr or unstimulated, were incubated with biotinylated Stat1 probe 2 from the GILT promoter. The complexes were immunoprecipitated with streptavidin–agarose, electrophoresed and probed for STAT1 by Western blotting. (b) STAT1 binding to γ-activated sequence element (GAS) sites in the GILT promoter is sequence-specific. Stat1−/− cells were transiently transfected with Stat1α, Stat1α-S727A and Stat1β-Y701F. The lysates were incubated with biotinylated Stat1 probe 2, and competition was carried out in the presence of a 50-fold excess of Stat1 consensus or p53 oligonucleotides. Proteins were immunoprecipitated and analysed for the presence of STAT1 by Western blotting.
Discussion
During an early immune response the expression of various immune molecules is induced. GILT is constitutively expressed in professional APCs and is also inducible in vitro in APCs by inflammatory cytokines such as IFN-γ, tumour necrosis factor-α (TNF-α) and interleukin-1β (IL-1β). Stat1 has been shown to regulate the IFN-γ-stimulated induction of GILT.12 However, we found that GILT is also constitutively expressed at detectable levels in other cell types not involved in antigen processing, such as mouse T cells and skin fibroblasts.9,10 Therefore, GILT is produced at basal levels without any extracellular stimuli. We were interested to determine whether Stat1 plays any role in the constitutive expression of GILT. We expected that the absence of Stat1 in Stat1−/− cells would reduce the expression of GILT. Surprisingly, the Stat1−/− mouse fibroblast cell line (MEF) showed increased levels of GILT protein, suggesting that STAT1 may exert a negative regulation on the constitutive expression of GILT.
Stat1 is a major transcription factor activated by IFN-γ and IFN-α/β signal transduction cascades that ultimately lead to the activation of antiviral, antiproliferative and immunomodulatory functions. However, it has also been shown that Stat1 is an active transcription factor involved in the constitutive, ligand-independent, transcription of some genes, such as caspase genes,24 and the LMP2 gene22,34, MHC class I.25 While ligand-induced, Stat1-mediated gene expression can either down-regulate or up-regulate the expression of target genes,22,25,35–37 most evidence suggests that the steady presence of STAT1 is necessary for constitutive expression of target genes, and hence the absence of Stat1 will lead to the down-regulation of gene expression. In this study we showed that STAT1 has a suppressive effect on the ligand-independent, constitutive activity of the GILT promoter.
In our experiments, the GILT promoter in Stat1−/− MEFs in the absence of stimulation with IFN showed a three- to fourfold increased activity of the firefly luciferase reporter gene when compared with WT MEFs. These findings are consistent with higher expression of the GILT protein in untreated Stat1−/− MEFs. However, upon treatment with IFN-γ, the levels of GILT protein do not increase in STAT1−/− MEFs, whereas GILT expression increases in WT MEFs, as expected. Therefore, STAT1 may play a dual role in the regulation of GILT expression: in the presence of inflammatory stimuli (e.g. IFNs) STAT1 rapidly increases the expression of GILT when it is necessary to process more antigens, whereas in the absence of inflammatory stimuli it is unnecessary for the cell to process more antigens and therefore not necessary to up-regulate the production of GILT.
Tyrosine phosphorylation in response to cytokine stimulation of cells is believed to be required for the nuclear translocation of cytoplasmic STAT1 proteins. However, it has been shown that phosphorylation of Y701 is not always necessary for the nuclear localization of STAT1.38,39 Phosphorylation of serine 727 occurs independently of phosphorylation of Y701 and it substantially enhances the transcriptional activity of STAT1.40 Here, we showed that phosphorylation of tyrosine and serine residues in STAT1 is not required for in vitro binding to putative GAS sites in the GILT promoter. We used STAT1 mutants that lack either S727 (Stat1α-S7272) or both Y701 and the C-terminus (Stat1β-Y701), required for transcriptional activation and interaction with CBP/p300 complex, for co-transfection with the firefly luciferase reporter gene, under the control of the GILT promoter, into Stat1−/− MEFs. Transfection of either mutant decreased the activity of the reporter gene to the level similar to that seen in WT cells. Therefore, our data suggest that neither phosphorylation of Y701 nor of the C-terminal portion of STAT1 is required for the constitutive suppression of the GILT promoter. However, the binding of STAT1 to putative GAS sites – either WT STAT1 or a phosphorylation mutant STAT1 – is specific and can be competed-out in a DAPA assay only by an oligonucleotide that contains a STAT1 consensus sequence.
STAT1 can exert its effect on target DNA either by direct binding or indirectly through the formation of complexes with other transcription factors. We hypothesized that the DNA-binding region of STAT1 may contain a site that is important for the constitutive interaction of STAT1 and the GILT promoter. Therefore, we tested whether known DNA-binding mutants – V426D/T427D,29 E428A/E429S30 and K544A/E545A,31– can alter the activity of the GILT promoter. Our luciferase reporter gene experiment indicated that only V426D/T427D was unable to decrease the activity of the GILT promoter, suggesting that STAT1 binding to DNA is necessary and that residues V426/T427 are the most important for the STAT1 suppressive effect on the ligand-independent activity of the GILT promoter. The mutant V426D/T427D is defective in the IFN-γ-induced STAT1 DNA binding to specific GAS sites and shows weakened, non-specific protein–DNA interactions,29 and therefore the implication is that GAS sites remain an important target for STAT1, even in the absence of IFN-γ stimulation. The DAPA confirmed that indeed the V426D/T427D (Mut 1) mutant cannot bind to GAS-like sites in the GILT promoter in vitro, whereas the K544A/E545A (Mut 3) mutant binds to GAS-like sites, albeit weakly. However, we were unable to show that the mutant E428A/E429S (Mut 2), which suppresses GILT promoter activity as in the WT, binds in vitro to a GAS-like site in the GILT promoter. This apparent discrepancy may be caused by very weak binding to the GAS site in the GILT promoter that is below the limits of detection by DAPA, and/or perhaps is caused by the binding of this mutant to another, as yet unidentified, transcription factor.
The fact that the absence of STAT1 increases the activity of the GILT promoter and GILT protein expression may be caused by competition/interaction of STAT1 with other transcription factors. For example, STAT3 can replace STAT1 in STAT1−/− cells to drive the transcription of certain genes in response to IFN-γ or interleukin-6.41 STAT1 and STAT3 dimers bind selectively to very similar, but not identical, elements27,42 and thus activate different, but overlapping, sets of genes. In addition, Egr-1 (also designated zif268, TIS 8, NFGI-A, Krox 24) has been identified as one of the transcription factors that targets GILT.43 Egr-1 is a member of the immediate-early gene family that includes FOS, JUN and early growth-response genes.44,45 Egr-1 binds to 5′-GCGGGGGCG-3′ consensus sequences within the promoter region of target genes.46 The GILT promoter contains several GC-rich domains in the vicinity of GAS-like sites and it is therefore possible that the binding of Egr-1 and STAT1 to some regions of the GILT promoter are mutually exclusive. The competition for binding to the GILT promoter, if any, remains to be shown.
In conclusion, although in most cases STAT1 plays a role in the induction of gene expression, it seems that its role is more complex and multifaceted. In mouse fibroblasts STAT1 appears to down-regulate the expression of genes not essential for cellular survival in a phosphorylation-independent manner. GAS or GAS-like sequences remain important targets for STAT1 binding to achieve this regulatory function.
Acknowledgements
This work was supported by American Heart Association Scientist development grant 0535032N awarded to M.M. We would like to express our gratitude to Dr M. Kaplan (Indiana University School of Medicine) for helpful input and to Dr D. Levy (NYU School of Medicine) for providing cell lines and plasmids as well as helpful suggestions.
Disclosures
The authors confirm that the manuscript, the title of which is given above, is original and has not been submitted elsewhere. Each author acknowledges that he/she has contributed in a substantial way to the work described in the manuscript and its preparation.




