Molecular cloning and expression analysis of a selenium-independent glutathione peroxidase identified from Manila clam Venerupis philippinarum
Abstract
Glutathione peroxidase (GPx) is a key enzyme of cellular detoxification systems that defend cells against reactive oxygen species. It can be divided into selenium-dependent GPx (Se-GPx) and selenium-independent GPx (non-Se-GPx) subfamilies. In this study, the full-length cDNA of a non-Se-GPx was identified from Venerupis philippinarum (denoted as VpGPx) using EST analysis and RACE approaches. The full-length cDNA of VpGPx encoded a polypeptide of 226 amino acids with the predicted molecular mass of 25.41 kDa. Phylogenetic analysis indicated that VpGPx had higher evolutional conservation to invertebrate than vertebrate counterparts and should be a new member of the GPx protein family. Spatial expression analysis found that VpGPx transcript was most abundantly expressed in hepatopancreas, gills and haemocytes, and weakly expressed in the tissues of mantle. After Vibrio anguillarum challenge, the expression of VpGPx transcript in overall haemocyte population was down-regulated in the first 6 h, and increased to the peak at 48 h. As time progressed, the expression level dropped to 0.5-fold of the control level at 96 h. All these results indicated that VpGPx was perhaps involved in the immune response against microbe infection and might be contributed to the clearance of bacterial pathogens.
Introduction
Manila clam, Venerupis philippinarum, is an important marine bivalve for commercial fisheries, accounting for about 80% of mudflat fishery production in China. However, with the rapid development of intensive culture and environmental deterioration, clams are continuously confronted with infectious microorganisms, which have resulted in enormous economic losses to the clam aquaculture (Munroe & McKinley 2007). Like other invertebrates, the absence of acquired immunity in clam makes them exclusively rely on the innate immune system to protect them against continuous threats from pathogens, especially in marine environments abundant in bacterial population (Austin 1988). Therefore, better understanding of the immune defense mechanisms of this species might be contributed to the development of management strategies for disease control and long-term sustainability of clam farming.
Phagocytosis of bacteria or bacterial products is usually coupled with the generation of reactive oxygen species (ROS) (Donati, Kantengwa & Polla 1991). ROS, including superoxide (O2−), hydrogen peroxide (H2O2), hydroxyl radicals (HO−) and singlet oxygen (1O2), are produced during the course of aerobic respiration and substrate oxidation (Dalton, Shertzer & Puga 1999). Although ROS play an important role in host defense, the excessive production of ROS can also cause metabolic malfunctions and damage to biological macromolecules, resulting in various diseases (Halliwell 1992; Matés, Pérez-Gómez & Núñez de 1999). To protect against ROS-generated oxidative stress, aerobic organisms have evolved complex non-enzymatic and enzymatic antioxidant defense systems (Matés et al. 1999; Matés, Pérez-Gómez & Blanca 2000). The main antioxidant enzymes include glutathione peroxidase (GPx, EC1.11.1.9), superoxide dismutase (SOD, EC1.15.1.1) and catalase (CAT, EC1.11.1.6) (Drevet 2006).
Among the enzymatic antioxidant system, GPxs constitute the second line of defense against lipid peroxidation, assuming an important role in the maintenance of the balance between ROS and antioxidants by catalysing the reduction of various organic hydroperoxide and hydrogen peroxidase using glutathione as hydrogen donor (Lawrence & Burk 1976; Ren, Huang, Akesson & Ladenstein 1997; Williamson & Bao 2000; Cossío-Bayúgar, Miranda & Holman 2005). Basically, GPxs can be divided into two types: selenium-dependent GPx (Se-GPx) and selenium-independent GPx (non-Se-GPx). Se-GPx has selenium-dependent glutathione peroxidase activity and contains selenocysteine (SeC) encoded by a TGA codon and can catalyse the reduction of organic and inorganic peroxides, whereas non-Se-GPx has no SeC and can only reduce organic peroxide by an entirely different mechanism (Tang, Gounaris, Griffiths & Selkirk 1995; Almar, Otero, Santos & Gonzalez Gallego 1998). To date, six mollusc Se-GPx from Unio tumidus (ABH10623), Dreissena polymorpha (ABP73388), Crassostrea gigas (ABS19600), Corbicula fluminea (ABQ24217), Haliotis discus discus (EF103379) and Chlamys farreri (EU183302) have been deposited in GenBank (Cossu, Doyotte, Jacquin, Babut, Exinger & Vasseur 1997; De Zoysa, Pushpamali, Oh, Whang, Kim & Lee 2008; Doyen, Bigot, Vasseur & Rodius 2008; Mu, Ni, Zhao, Wang, Song, Li, Zhang, Qiu & Cong 2010). Moreover, scallop Se-GPx has been proved responsive against Vibrio challenge and oxidative stress (Mu et al. 2010), indicating that Se-GPx was potentially involved in the immune response.
To our knowledge, genes encoding homologues of non-Se-GPx have been isolated from Escherichia coli (Friedrich, DeVeaus & Kadner 1986), plants (Criqui, Jamet, Parmentier, Marbach, Durr & Fleck 1992; Holland, Ben-Hayyim, Faltin, Camoin, Strosberg & Eshdat 1993) and mammals. However, only two non-Se-GPxs was identified from mollusc Aplysia californica (AF510851) and Crassostrea gigas (CAK22382) so far (Doyen et al. 2008). The main objectives of the present study were to clone the full-length cDNA of non-Se-GPx from V. philippinarum, and to investigate the tissue-specific expression and temporal expression profile of VpGPx transcript post pathogen challenge, and hopefully lay the foundation for better understanding of VpGPx involved in the immune response of clam.
Materials and methods
Animals and bacterial challenge
The clams V. philippinarum (average weight 9 ± 2 g) were purchased from a local farm, and acclimated for a week before commencement of the experiment. After the acclimation period, the clams were randomly divided into six tanks with 50 L capacity, each containing 50 individuals. The temperature was held at 20–22°C throughout the whole experiment. The salinity for the supplied seawater was kept at 30‰.
For the Vibrio anguillarum challenge experiment, one tank served as the control group. The clams in the other five tanks were immersed with high density of V. anguillarum with final concentration of 107 CFU mL−1. The infected clams were randomly sampled at 6, 12, 24, 48, 72 and 96 h respectively. The haemocytes from the control and infected groups were collected using a syringe and centrifuged at 2000 g, 4°C for 10 min to harvest the haemocytes. Five individuals were randomly selected from different tanks as replicates for each time point. The haemocyte pellets were immediately subjected to RNA extraction and qPCR analysis individually.
RNA isolation and cDNA synthesis
Total RNA was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The cDNA first-strand synthesis was carried out based on M-MLV RT Usage information (Promega, Shanghai, China) using RQ 1 DNase (Promega)-treated total RNA as template. cDNA mix was diluted to 1:5 and stored at −80°C for subsequent expression analysis.
Cloning the full-length cDNA of VpGPx gene
A cDNA library was constructed with the haemocytes of manila clam as previously described (Li, Qiu, Ning, Chen, Qin, Wu & Zhao 2010). BLAST analysis of the EST sequences revealed that an EST of 375 bp was highly similar to GPx identified previously, and this EST sequence was selected for further cloning. Four specific primers, sense primers P1 and P2 and reverse primers P3 and P4 (Table 1) were designed based on the EST to clone the full-length cDNA of VpGPx. The nested PCR strategy was applied to clone the 3′ end of VpGPx using sense primer P1, P2 and reverse primer T7, whereas sense primer oligo(dG)-adaptor and reverse primer P3, P4 were used to get the 5′ end of VpGPx. The PCR products were cloned into pMD18-T simple vector (TaKaRa, Dalian, China) and sequenced bi-directionally with primers M13-47 and RV-M (Table 1). The sequencing results were verified and subjected to cluster analysis.
| Primers | Sequence(5′–3′) | Sequence information |
|---|---|---|
| P1 | TCAAAGGGACTTCCACTC | 3′-RACE primer |
| P2 | TAGACCCAGTGGAGAAGG | 3′-RACE primer |
| T7 | GTAATACGACTCACTATAGGGC | 3′-RACE primer |
| P3 | CTTCTCCACTGGGTCTAAC | 5′-RACE primer |
| P4 | AGTCTGGACCAACAATAAA | 5′-RACE primer |
| Oligo(dG)-adaptor | GGCACGCGTCGACTAGTACG10 | 5′-RACE primer |
| P5 | GACGGATGGGCAATCTTGTTCTC | Real-time VpGPx primer |
| P6 | AGGTGAGTGGAAGTCCCTTTGAG | Real-time VpGPx primer |
| P7 | CTCCCTTGAGAAGAGCTACGA | Real-time actin primer |
| P8 | GATACCAGCAGATTCCATACCC | Real-time actin primer |
| M13-47 | CGCCAGGGTTTTCCAGTCACGAC | Sequencing primer |
| RV-M | GAGCGGATAACAATTTCACACAGG | Sequencing primer |
Sequence analysis
The searches for nucleotide and protein sequences similarities were conducted using BLAST algorithm at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast). The deduced amino acid sequence of VpGPx was analysed with the Expert Protein Analysis System (http://www.expasy.org/). signalp 3.0 program was utilized to predict the presence and location of signal peptide (http://www.cbs.dtu.dk/services/SignalP/). Multiple alignment of VpGPx with GPxs from other organisms was performed with the ClustalW Multiple Alignment program (http://www.ebi.ac.uk/clustalw/) and Multiple Alignment show program (http://bio-soft.net/sms/). An unrooted phylogenetic tree was constructed based on the sequence alignment by the neighbour-joining (NJ) algorithm embedded in mega 3.1 program (http://www.megasoftware.net/). To derive the confidence value for the phylogeny analysis, bootstrap trials were replicated 1000 times.
VpGPx mRNA expression in different tissues and the response to bacterial challenge
The mRNA expression of VpGPx in different tissues and the response to bacterial challenge was measured using quantitative real-time PCR. Total RNA were extracted from different tissues of five individuals, including muscle, gill, haemocyte, hepatopancreas and mantle. A 292-bp product was amplified with VpGPx gene-specific primers P5 and P6 (Table 1) from cDNA mix, and the PCR product was sequenced to verify the PCR specificity. Two β-actin primers, P7 and P8 (Table 1) were used to amplify a 121-bp fragment as internal control.
Real-time PCR amplification was carried out in an Applied system 7500 fast real-time PCR system. The amplifications were performed in triplicates in a 25-μL reaction volume containing 12.5 μL of 2× SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA), 1 μL (each) of forward primer and reverse primer (5 μmol L−1), 1 μL of 1:5 diluted cDNA and 9.5 μL of PCR grade water. The thermal profile for real-time PCR was 50°C for 2 min and 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 59°C for 1 min. All analyses were based on the CT values of the PCR products. The CT was defined as the PCR cycle at which the fluorescence signal crossed a threshold line that was placed in the exponential phase of the amplification curve. Dissociation curve analysis of amplification products was performed at the end of each PCR reaction to confirm that only one PCR product was amplified and detected. After the PCR program, data were analysed using ABI 7500 sds software V2.01. Real-time PCR efficiency for the targets was calculated from the slopes of the standard curve (CT vs. log concentration) using the formula E = 10[−1/slope] (Pfaffl, Horgan & Dempfle 2002). Both targets amplified with high efficiency during real-time PCR: β-actin (1.94) and VpGPx (1.87). To maintain consistency, the baseline was set automatically by the software. The 2−ΔΔCT method was used to analyse the expression level of VpGPx (Livak & Schmittgen 2001). All data were given in terms of relative mRNA expressed as mean ± SE (n = 5). The data were then subjected to one-way analysis of variance (anova) followed by an unpaired, two-tailed t-test. Difference was considered to be significant at P < 0.05.
Results and discussion
Cloning and structural analysis of VpGPx
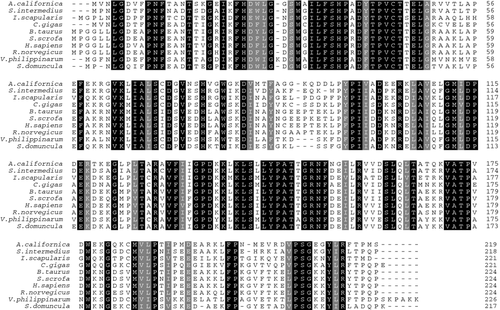
The full sequence of VpGPx cDNA obtained from our study consisted of 1049 bp (GenBank accession no. GQ384395), encoding a polypeptide of 226 amino acids with the predicted molecular mass of 25.41 kDa (Fig. 1). Signal P analysis revealed that VpGPx was a non-Se-GPx, for no signal peptide was identified in the deduced amino acid sequence. ClustalW analysis revealed that the deduced amino acid sequence of VpGPx displayed high identity with other reported GPxs (Fig. 2). For example, VpGPx shared 69% identity to GPx from A. californica (AF510851) and Suberites domuncula (CAC38779), 64% identity to Strongylocentrotus intermedius (ADK11694), 61% identity to Ixodes scapularis (AAK97814), 60% identity to C. gigas (CAK22382) and 59% identity to Rattus norvegicus (NP_446028) and Homo sapiens (NP_004896).

Phylogenetic analysis of VpGPx
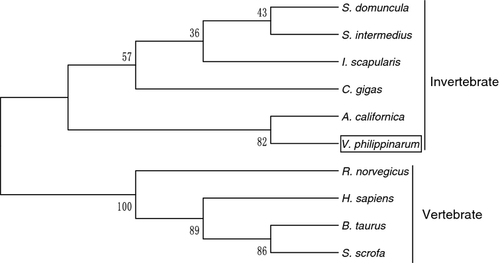
To evaluate the molecular evolutionary relationships of VpGPx against other GPxs, a phylogenetic tree was constructed based on the protein sequences using the NJ method (Fig. 3). According to the phylogenetic tree, the GPx members were mainly clustered into two groups by their invertebrate and vertebrate origin. VpGPx was first clustered with GPx from mollusc A. californica, then formed a sister group with those from invertebrates, and further grouped with those from vertebrates. The relationships displayed in the phylogenic tree were in good agreement with traditional taxonomy, suggesting that this protein family might have a primarily similar functional role.
Tissues distribution of VpGPx
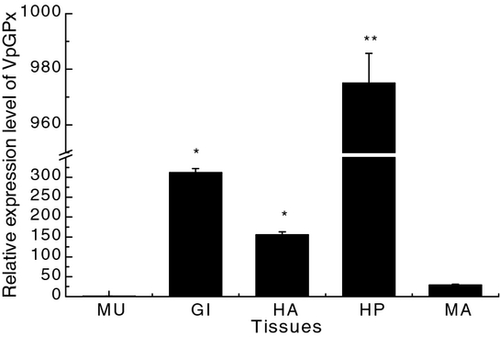
To examine the tissue distribution profile of VpGPx, total RNA was isolated from hepatopancreas, gill, muscle, haemocyte and mantle. The expression of VpGPx transcript was predominantly detectable in hepatopancreas, gills, haemocytes, and to a less degree in the tissue of mantle (Fig. 4). Similar tissue expression pattern was also observed in Se-GPx of H. discus discus, U. tumidus, C. farreri and Litopenaeus vannamei (Doyen, Vasseur & Rodius 2006; Liu, Tseng & Cheng 2007; De Zoysa et al. 2008; Mu et al. 2010). The highest expression of VpGPx in hepatopancreas reasonably supported the fact that hepatopancreas was the main metabolic center for ROS production in mollusc (Bianchini & Monserrat 2007). VpGPx mRNA was also found highly expressed in gills, indicating that more VpGPx was required to protect the tissue from damage of ROS generated due to high oxygen pressure. Moreover, the high expression of VpGPx in haemocytes suggested the potential immune functions of this protein, as haemocytes were crucial to immunity defense not only by direct sequestration and killing of foreign invaders but also by synthesis and exocytosis of bioactive molecules (Hoffmann, Kafatos, Janeway & Ezekowitz 1999; Roch 1999; Tincu & Taylor 2004).
VpGPx expression pattern in response to bacterial challenge
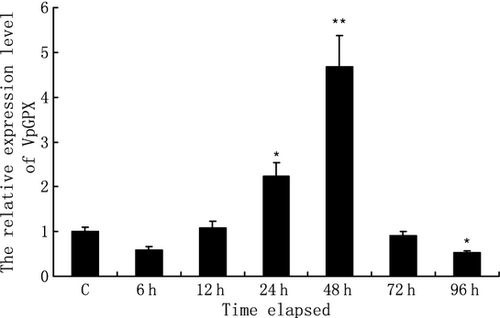
The mRNA expression level of VpGPx in haemocytes of clams after bacterial challenge was quantified using real-time RT-PCR with β-actin as internal control (Fig. 5). During the first 6 h, the expression of VpGPx decreased down to 50% of the control. As time progressed, the expression level of VpGPx was up-regulated gradually and reached 4.7-fold of the control group at 48 h. After that, the expression level was sharply decreased. At 96 h, VpGPx mRNA level dropped greatly and was only 0.5-fold compared with the control. One-way anova showed statistically significant difference in VpGPx gene expression at 24 h (P < 0.05), 48 h (P < 0.01) and 96 h (P < 0.05) post-infection.
As one kind of major antioxidant enzyme to eliminate H2O2, GPx can convert H2O2 to water before hydroxyl radicals can be produced (Aruoma 1998). In the previous studies, L.vannamei GPx activity and its mRNA transcription was increased significantly to eliminate the excess of H2O2 (Liu et al. 2007). This phenomenon of antioxidant defense against H2O2 after pathogen infection by increasing GPx activity was also found in disc abalone, H. discus discus (De Zoysa et al. 2008). In the present study, during the early phase of Vibrio infection, the expression of VpGPx was restricted at a lower level, perhaps to avoid the degradation of ROS produced to kill invaded pathogens. As time progressed, the adverse effect of ROS is gradually enhanced compared with the invasion of the microorganism, resulting in the up-regulated expression of VpGPx to minimize the negative effects. The up-regulation of VpGPx mRNA in haemocytes may be beneficial for host to mitigate the damage resulted from high amount of ROS induced by bacterial challenge. Similarly, Mu et al. (2010) have shown up-regulated GPx3 transcripts in C. farreri haemocytes upon bacteria stimulation. These results collectively suggested that GPx were involved in the immune response in these species, and further highlighted the functional importance of VpGPx in the clam immune system.
Acknowledgments
The project was supported by grant from NSFC (30901115), and in part by the 100 Talents Program of the Chinese Academy of Sciences and Key Laboratory of Marine Spill Oil Identification and Damage Assessment Technology (201115), SOA.




