The first identification of molluscan Ecsit in the Pacific oyster, Crassostrea gigas, and its expression against bacterial challenge
Abstract
The evolutionarily conserved signalling intermediate in Toll pathways (Ecsit) plays crucial roles in innate immunity, embryogenesis and mitochondria-related functions. In this study, the first molluscan Ecsit gene (named as CgEcsit) is isolated from Crassostrea gigas based on the expressed sequence tags-based rapid amplification of cDNA ends polymerase chain reaction (PCR) method. The cloned cDNA of CgEcsit was 1545 bp in length, containing a 1356 bp open reading frame encoding a 451 amino acid protein. Sequence comparison showed that the deduced amino acid sequence of CgEcsit shared approximately 27–33% identities with other known Ecsits from invertebrates and vertebrates. The genomic DNA of CgEcsit covered a length of approximately 6 kb and consisted of six exons and five introns, which represented a closer genomic architecture with Ecsits from vertebrates than that of model invertebrates. The expression pattern of CgEcsit was analysed by real-time quantitative reverse transcript-PCR, in various tissues and especially in haemocytes under vibrio challenge stress. The expression in the haemolymph was challenged dramatically by the bacteria Vibrio anguillarum. These results suggested that Ecsit in C. gigas should be a potential factor of the oyster defence system, and the function and mechanism of CgEcsit might contribute to better disease management strategies of oyster farming.
Introduction
The evolutionarily conserved signalling intermediate in Toll pathways (Ecsit) was firstly reported in 1999 to be an intermediate in Toll-like receptor (TLR) signalling pathways and a bridge between TRAF6 and MEKK-1 (Kopp, Medzhitov, Carothers, Xiao, Douglas, Janeway & Ghosh 1999). In 2003, Ecsit was also reported to bind to the promoters of specific bone morphogenetic proteins (Bmp) target genes, to interact with Smad1 and Smad4 and to play critical roles during mouse embryogenesis (Xiao, Shim, Kluppel, Zhang, Dong, Flavell, Fu, Wrana, Hogan & Ghosh 2003). Vogel, Janssen, van den Brand, Dieteren, Verkaart, Koopman, Willems, Pluk, van den Heuvel, Smeitink and Nijtmans (2007) reported that Ecsit also localized in the mitochondria, where it interacted with the molecular chaperone NDUFAF1 and functioned as a fundamental component required by the efficient assembly of mitochondrial NADH:ubiquinone oxidoreductase. Both the TLR pathway and the Bmp pathway are important in organism vital movement, and they are of great significance in cell growth, division, migration and apoptosis (Takeda & Akira 2004; Haynes, Gutierrez, Aycinena, Tsonis & Del Rio-Tsonis 2007; Frandsen, Gunn, Muratoglu, Fossett & Newfeld 2008).
The Pacific oyster, Crassostrea gigas (Thunberg 1793), is native to Japan, and has been introduced to all continents except Antarctica (Ucko, Colorni, Kvitt, Diamant, Zlotkin & Knibb 2002; Guo 2009). C. gigas is important in aquaculture, and had the highest annual production among the freshwater and marine organisms (Hedgecock, Gaffney, Goulletquer, Guo, Reece & Warr 2005). Despite the high production, oyster cultivation has suffered drastically from high mortality in the recent years due to a complex interaction of over-harvesting, anthropogenic impaction, physical factors (salinity and temperature) and disease (pathogeny infestation). A large-scale epidemiological study in bacteria pathogenic to the Pacific oyster in France was conducted and it was found that Vibrio was the major reason for the severe production loss in the oyster industry (Saulnier, De Decker & Haffner 2009; Saulnier, De Decker, Haffner, Cobret, Robert & Garcia 2010). Besides, oysters are sometimes bacterial carriers and closely related to human health. Recently, the economic and ecological importance of oysters has made them a model in pathogen stress defence studies (Hedgecock et al. 2005). Understanding the immune defence mechanisms of oysters to pathogenic bacteria would contribute to disease management strategies and the development of sustainable oyster culture.
Ecsit is multifunctional and is the unique intermediate point between the TLR pathway and the Bmp pathway reported to date. However, before this study, there was no Ecsit identified in the molluscs. Our specific goals were (1) to clone Ecsit from the C. gigas, the first in molluscan, and to compare its nucleotide sequence similarity with other Ecsits; (2) to determine the genomic structure of CgEcsit and to compare it with other known Ecsit genes; and (3) to study the expression dynamic of CgEcsit in different tissues and its temporal expression in haemocytes after Vibrio anguillarum treatment.
Material and methods
Animals, bacterial challenge and tissue collection
Live healthy oysters, average 110 mm in shell height, were purchased from Qingdao, China, and acclimatized in seawater tanks at a temperature of 18 ± 1.0 °C throughout the experiments. V. anguillarum (strain no. MVM425) was suspended in a phosphate buffer solution (PBS, pH7.2). The concentration of bacterial culture was calculated by the optical density at 550 nm (1 unit OD 550 equals to 5 × 108 bacteria mL−1). Oysters were divided into two groups and each group consisted of 50 individuals. By filing the shell, oysters were injected into the adductor muscle with either 100 μL V. anguillarum or 100 μL PBS. At 0, 3, 6, 12, 24 and 48 h after being challenged, three oysters were randomly sampled in each subgroup, according to Itoh and Takahashi (2009). The haemolymph was collected from the pericardial cavity using 1 mL sterile syringes, and was then centrifuged at 1000 g for 10 min, 4 °C according to Gueguen, Cadoret, Flament, Barreau-Roumiguiere, Girardot, Garnier, Hoareau, Bachere and Escoubas (2003). Other tissues were surgically obtained and also three samples in each subgroup.
Cloning cDNA and gDNA of CgEcsit
Total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA), and DNaseI (Promega, USA) treatment was conducted to eliminate contaminating of genomic DNA. Reverse transcription reactions were performed using M-MLV reverse transcriptase (Promega, Madison, WI, USA). To verify the sequence of the CgEcsit expressed sequence tags (ESTs) (GenBank accession number: M858613, BQ427193, AM865338, AM865331, AM865335), we designed a pair of primers consisting of a forward primer ECSITF1 and a reverse primer ECSITR1 (Table 1). 3′-ends rapid amplification of cDNA end was carried out using the gene-specific primer ECSITGSP1 and the Oligo(dT)-adaptor in the first-round polymerase chain reaction (PCR) and ECSITGSP2 and the anchor primer in the second-round PCR (Invitrogen). The genomic DNA was extracted from the gills using the Phenol–Chloroform method. Primers (ECSITF1, ECSITgR1), (ECSITgF2, ECSITgR2) and (ECSITgF3, ECSITgR3) were used to amplify CgEcsit genomic DNA. All the purified PCR products were cloned into the pMD18-T vector (TaKaRa, BIO INC., Shiga, Japan) and sequenced in both directions. The resulting sequences were verified and subjected to comparative analysis. The primers used in this study are shown in Table 1.
| Name | Sequence | Function |
|---|---|---|
| ECSITF1 | TGTGATASGGAACAYCCTTGGTG | Cloning primer of cDNA and gDNA |
| ECSITR1 | ACCTTGGAGCGTTGGGAGAAT | Cloning primer of cDNA |
| ECSIT_3CF1 | CAGGATGCCGCTCTGAGAGTTCTGACA | 3′RACE PCR |
| ECSIT_3CF2 | CASATTCTCCCAACGCTCCAAGGT | 3′RACE PCR |
| Oligo(dT)-adaptor | GGCCACGCGTCGACTAGTACT16 | 3′RACE adaptor primer |
| AP | GGCCACGCGTCGACTAGTAC | RACE adaptor primer |
| ECSITgR1 | ATACCAGGACATGGCGAGCAT | Cloning primer of gDNA |
| ECSITgF2 | AGATGCTCGCCATGTCCTGST | Cloning primer of gDNA |
| ESCITgR2 | GCACGTACAGTCCAGCATTCAACT | Cloning primer of gDNA |
| ESCITgR3 | GGGAACCTCACCAAATTTATTCTG | Cloning primer of gDNA |
| ESCITgF3 | CCTACATATGAAAGTACAACCGAC | Cloning primer of gDNA |
| ESCITRTF | CCCTCCCTCCAAACCAATCA | qRT-PCR |
| ESCITRTR | AGTCCCGTCCTCCTGCTCAT | qRT-PCR |
| EFF-real | AGTCACCAAGGCTGCACAGAAAG | qRT-PCR |
| EFR-real | TCCGACGTATTTCTTTGCGATGT | qRT-PCR |
- RACE, rapid amplification of cDNA ends; qRT-PCR, real-time reverse transcript polymerase chain reaction.
Analysis of nucleotide and deduced amino acid sequences of CgEcsit
Expressed sequence tags homologous to Ecsit were searched in the NCBI database using the blastn package. cDNA and deduced amino acid sequences were analysed using dnaman software. Deduced protein domains were predicted in the SMART online analysis (http://smart.embl-heidelberg.de/). Calculated molecular mass and theoretical isoelectric points were predicted using Protein MolWt &AA Composition Calculator (http://www.proteomics.com.cn/proteomics/pi_tool.asp). The cloned CgEcsit was identified as cytoplasmic protein by Cello (http://cello.life.nctu.edu.tw/), the mitochondrial targeting sequence was predicted by TargetP (http://www.cbs.dtu.dk/services/TargetP/) and the nuclear subcellular localization sequence was predicted by Subloc V1.0 (http://www.bioinfo.tsinghua.edu.cn/SubLoc/cgi-bin/eu_subloc.cgi). The GC content was calculated using CodonW (http://codonw.sourceforge.net/). The intron–exon structure was estimated using the local blastn package. Multiple sequence alignment of Ecsit protein sequences was created using ClustalX 1.83 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). A phylogenetic tree of the selected Ecsit proteins was constructed using the neighbour-joining method, and the reliability of the branching was tested using bootstrap resampling (with 1000 pseudo-replicates). The figure was drawn using the MEGA soft package 4.0.
Quantitative analysis of CgEcist transcription
A real-time reverse transcript polymerase chain reaction (qRT-PCR) was carried out using an ABI 7500 fast Real time Thermal Cycler according to the manual (Applied Biosystems, Foster City, CA, USA). SYBR Green I Premix ExTaqTM (TaKaRa) was used as a fluorescence dye. In qRT-PCR, reverse transcription was conducted with the OligodT18 primer and the random hexamer primer (N6) mixed in proportion. Two primers ESCITRTF and ESCITRTR (Table 1) were designed to amplify the 244 base pairs (bp) fragment of CgEcsit, with the EF gene as an internal control (primers EFF-real and EFR-real). Cycling conditions were 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 20 s and 72 °C for 30 s and then a melt curve stage after the cycling stage. The specificity of qRT-PCR products was analysed by agarose gel electrophoresis and melting curve analysis. In the tissue expression experiment, the gonad was used as a reference sample (called the calibrator), while time 0 h was used as the calibrator in the challenge experiment. The ΔCt for each sample was subtracted from the ΔCt of the calibrator; the difference was called the ΔΔCt value. The expression level of CgEcist could be calculated by 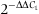 .
.
Statistical analysis
The data obtained were subjected to student's t-test to determine the difference in the mean values between the controls and the treatments. Differences were considered to be significant at P<0.05. All data were presented in terms of relative mRNA expressed as mean ± SE (n=5). Statistical analysis was performed using software spss 13.0.
Results
cDNA cloning and sequence analysis of CgEcsit
cDNA of an Ecsit with a complete open reading frame (ORF) was acquired from C. gigas and designated as CgEcsit (GenBank accession number HQ225834) (Fig. 1). The length of CgEcsit cloned was 1545 bp, containing a 5′untranslated regions (UTR) of 93 bp, a 1356 bp ORF that encoded a 451 amino acid residue protein, a 3′UTR of 82 bp that included a stop codon TAA, a putative polyadenylation signal (AATAAA) and a Ploy(A) tail. CgEcsit involved an adenine three bases upstream of the start codon ATG and another adenine, followed by the start codon ATG, which followed the kozak rule. Besides, four stop codons (TGA, TGA, TAA, TAA) occurred upstream of the first ATG with the same coding frame of CgEcsit. These two aspects above suggested that ATG (94–96) in CgEcsit was the initiation methionine of the translation.

Nucleotide and the deduced amino acid sequences of CgEcsit. The start codon (ATG), the stop codon (TAA) and the polyadenylation signal sequence (ATTAAA) are in bold. *Indicates the translation stop codon.
The deduced CgEcsit protein is shown in 1, 2. A classical ECSIT domain was annotated from amino acids location 91 to 303. Besides, CgEcsit was identified as a cytoplasmic protein, while it had both the mitochondrial targeting sequence and the nuclear subcellular localization sequence. The calculated molecular mass of the deduced CgEcsit was 52.78 kDa, and the theoretical isoelectric point was 6.11. CgEcsit was the first bivalve Ecsit reported with a complete ORF.
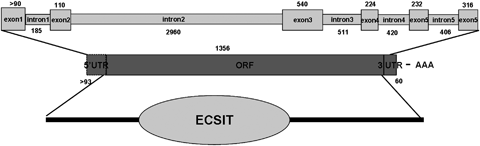
Schematic representation of the CgEcsit gene structure. Exons and introns are represented by boxes and lines. Numbers above or below the diagrams show length in base pairs. ORF, open reading frame; UTR, untranslated regions.
Genomic sequence of CgEcsit
The cloned genomic sequence of CgEcsit contained 5994 nucleotides (GenBank accession number HQ225835), which consisted of six exons and five introns (Fig. 2). The first intron located within 5′UTR, and other introns analysed in this study were present at the ORF, and all the intron splicing sites followed the GT/AG rule. Exons were calculated with a much higher GC content than introns. The GC contents in six exons were estimated as 37.2%, 40.7%, 38.3%, 52.7%, 49.8% and 45.5%, respectively, while that of five introns were estimated as 28.1%, 36.9%, 27.4%, 29.5% and 33.1%.
The genomic architecture of the coding sequences of CgEcsit was compared with those of other selected species (Table 2). Although Ecsit protein sequences had a similar length from 324 amino acids (Nematostella vectensis) to 452 amino acids (Danio rerio), the genomic length of the Ecsit genes was identified to show a huge difference, from 1192 nucleotides (Caenorhabditis elegans) to 12 913 nucleotides (Homo sapiens). C. gigas took the leading position in terms of amino acid sequence length (451 amino acids) in invertebrates, and the length in other invertebrates was all <400 amino acids. Genome organizations of Ecsit genes were also varied in the selected species. Table 2 shows that seven exons were present at the genomic sequences of Ecsit in vertebrate, five in Strongylocentrotus purpuratu and C. gigas, four in C. elegans, three in N. vectensis and only one in Drosophila melanogaster.
| Species | E1 | I1 | E2 | I2 | E3 | I3 | E4 | I4 | E5 | I5 | E6 | I6 | E7 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Homo sapiens | 93 | 4800 | 420 | 524 | 222 | 5007 | 60 | 140 | 150 | 139 | 105 | 1028 | 243 |
| Mus musculus | 93 | 1502 | 420 | 1413 | 222 | 850 | 60 | 98 | 150 | 338 | 105 | 401 | 255 |
| Bos taurus | 93 | 2399 | 420 | 222 | 222 | 511 | 60 | 133 | 150 | 160 | 105 | 567 | 249 |
| Danio rerio | 138 | 400 | 378 | 119 | 222 | 2503 | 60 | 328 | 150 | 549 | 120 | 3813 | 252 |
| Strongylocentrotus purpuratu | 111 | 622 | 399 | 631 | 222 | 1008 | 242 | 933 | 219 | ||||
| Drosophila melanogaster | 1227 | ||||||||||||
| Caenorhabditis elegans | 153 | 44 | 228 | 48 | 498 | 50 | 171 | ||||||
| Crassostrea gigas | 107 | 2960 | 536 | 515 | 224 | 420 | 232 | 406 | 254 | ||||
| Nematostella vectensis | 630 | 582 | 208 | 435 | 137 |
- E, Exon; I, Intron.
Homology and phylogenetic analysis of CgEcsit
Multiple alignment of CgEcsit with other selected Ecsits from the NCBI database suggested that the primary amino acid structures of the Ecsit genes were relatively conserved (Fig. 3). CgEcsit shared high similarity to other previously reported Ecsits, even Ecsits of vertebrate, such as 33% identities to D. melanogaster, 32% identities to Xenopus laevis, 31% identities to S. purpuratu and H. sapiens, 30% identities to Bos Taurus, Mus musculus and C. elegans, 29% identity to D. rerio and finally 27% identity to N. vectensis. Furthermore, a phylogenic tree was conducted based on the amino acid sequences of selected species based on the neighbour-joining method (Table 3 and Fig. 4). All the deuterostome clustered together as a subgroup, and the protostome clustered into another subgroup, except N. vectensis. In the protostome subgroup, the CgEcsit first clustered with CeEcsit and then with DmEcsit. The topology approximately reflected the taxonomic classification of the corresponding animals.
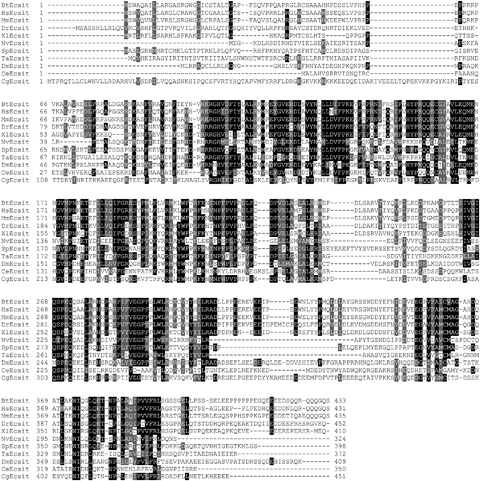
Multiple alignment of CgEcsit with other Ecsits deposited in GenBank. The black shadow region indicates the positions where all sequences share the same amino acid residue. Gaps are indicated by dashes to improve the alignment. The species and the GenBank accession numbers are given in Table 3.
| Species | GenBank no | Abbreviation |
|---|---|---|
| Crassostrea gigas | HQ225834 | CgEcsit |
| Nematostella vectensis | EDO47956 | NvEcsit |
| Caenorhabditis elegans | AAF60446 | CeEcsit |
| Drosophila melanogaster | AAL48530 | DmEcsit |
| Strongylocentrotus purpuratu | XM_789351 | SpEcsit |
| Xenopus laevis | AAI29632 | XlEcsit |
| Danio rerio | AAI24209 | DrEcsit |
| Mus musculus | AAF01219 | MmEcsit |
| Bos Taurus | AAI04573 | BtEcsit |
| Homo sapiens | AAF6210 | HsEcsit |

Neighbour-joining tree of full-length Ecsit proteins with 1000 bootstrap trials by the mega program. The scale bar indicates a branch length of 0.2, and bootstrapping values are indicated at the fork. Data obtained from GenBank are given in Table 3.
The distribution of CgEcsit in different tissues
Quantitative RT-PCR was performed to investigate the distribution of CgEcsit mRNA in different tissues. The CgEcsit transcript was detected in all the tested tissues of C. gigas, but the expression levels were considerably different (Fig. 5). The LSD t-test showed that the expression levels in the haemolymph, digestive gland and muscle were significantly higher than those in the gill, mantle and gonad. The haemolymph was identified to have the highest CgEcsit expression level.
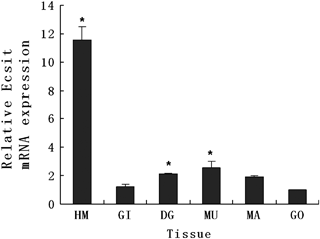
Quantitative real-time PCR analysis of the CgEcsit gene expression in various tissues of the Pacific oyster, Crassostrea gigas. Elongation factor gene expression was used as an internal control. Vertical bars represent the mean ± SD (N=3). *P<0.05 between gills and other tissues. HM, haemolymph; GI, gill; DG, digestive gland; MU, muscle; MA, mantle; GO, gonad.
Expression of CgEcsit mRNA in the haemolymph after bacterial challenge
To determine the expression pattern of CgEcsit in the haemolymph after bacterial challenge, two groups that had been challenged with PBS (control) and V. anguillarum were selected and qRT-PCR as established at post-challenge (0, 3, 6, 12, 24 and 48 h) (Fig. 6). The expression level of CgEcsit increased to 1.26-fold at 3 h post-injection, and slightly increased to 1.93-fold at 12 h, then rapidly reaching the highest level to 6.42-fold at 24 h, and finally recovering to 1.67-fold of the origin level at 48 h post-challenge. Significant differences between oysters injected with PBS and with V. anguillarum were observed at 3, 6, 12, 24 and 48 h after the challenge (P<0.05).
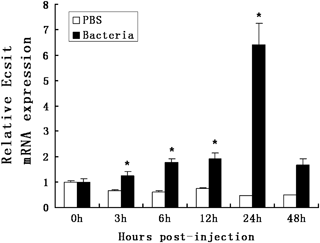
CgEcsit mRNA expression in oyster haemolymph following a V. anguillarum challenge. Elongation factor (EF) gene expression was used as an internal control. Vertical bars represent the mean ± SD (N=3). White indicates injected with PBS (negative control) and black Vibrio anguillarum. Results that are significantly different (P<0.05) between challenged and control at the same time point are shown with an asterisk.
Discussion
The Lophotrochozoa (spiralian and lophophorate protostomes) are the major members of the general evolutionary process; however, few studies have focused on this lineage compared with the other clade of Protostomes, the Ecdysozoa. Molluscs (Bilateria, Lophotrochozoa) are the second most diverse group of animals, with about 93 000 extant species; the focus of this study of the Lophotrochozoa is especially on the genetic map (Hubert & Hedgecock 2004), transcriptomics (Hedgecock, Lin, DeCola, Haudenschild, Meyer, Manahan & Bowen 2007) and BAC library construction (Cunningham, Hikima, Jenny, Chapman, Fang, Saski, Lundqvist, Wing, Cupit, Gross, Warr & Tomkins 2006).
Ecsit has been reported to play an essential role in innate immunity, embryogenesis and assembly or the stability of the mitochondrial complex I (Kopp et al. 1999; Xiao et al. 2003; Vogel et al. 2007); however, no mollucan Ecsit had been identified so far. In the present study, an Ecsit gene with a complete ORF from C. gigas (CgEcsit) was cloned. Deduced amino acids of CgEcsit were predicted with mitochondrial targeting and nuclear subcellular localization, which suggested the potential role of CgEcsit in the stability of mitochondrial complex I as reported in HEK293 cells. The CgEcsit gene was composed of six exons and five introns; the GC contents of six introns were between 28.1% and 33.1%, while the GC contents of exons were all higher than 37%. Generally, the GC content of CgEcist was similar to the overall GC pattern of the C. gigas genome (Hedgecock et al. 2005). A significant difference in the GC content between introns and exons illustrated that exons might undergo stronger evolutionary pressure than introns, because a higher GC content is always behalf of more important function and purifying selection (Powell & Moriyama 1997).
Earlier studies report that a diversity of immune genes in C. gigas are closer to those of vertebrates than to those of ecdysozoans (Zhang, Li & Zhang 2011a, b). A comparative study of amino acid similarity, genomic arrangement and phylogenetic tree structures was conducted to investigate the evolutionary lineage of CgEcsit. Although the amino sequences of CgEcsit showed the highest identities to D. melanogaster, its length was much closer to that of vertebrates. The genome architecture of Ecsit in the vertebrate was highly conserved, especially in terms of exon arrangement. In contrast, the exon–intron structure in invertebrates was looser. The genomic sequence of Ecsit consisted of 5 exons in C. gigas and S. purpuratu, but only one exon in D. melanogaster. It is reported that there had been extensive intron gain and/or intron loss during evolution in eukaryote genomes (Jeffares, Mourier & Penny 2006). The result that lower intron density in the Ecsit gene of D. melanogaster was in accordance with the earlier studies that fly has undergone extensive intron loss (Jeffares et al. 2006).
The deduced protein of CgEcsit showed high-level similarities to selected Ecsits reported in other species. The Neighbour-Joining tree divided Ecsits into two parts: the protostome and the deuterostome. The exception was NvEcsit from N. vectensis; it belongs to the protostome but was part of the deuterostome subgroup. This phenomenon has been elaborated at the level of the N. vectensis genome sequence. The sea anemone genome has a gene repertoire, exon–intron structure and large-scale gene linkage more similar to vertebrates than to flies or nematodes (Hemmrich, Miller & Bosch 2007; Putnam, Srivastava, Hellsten, Dirks, Chapman, Salamov, Terry, Shapiro, Lindquist, Kapitonov, Jurka, Genikhovich, Grigoriev, Lucas, Steele, Finnerty, Technau, Martindale & Rokhsar 2007).
Ecsit was reported to interact with TRAF6 and then play a significant role in the TLR signalling pathway. Hametocytes are effective cells in immune responses of mollusks, which participate directly in pathogen elimination by phagocytes and encapsulation and also produce humoral components including lysosomal emzymes, aminopeptidases, lectins and antimicrobial molecules (Matozzo, Rova & Marin 2007). The highest expression in the haemolymph suggested a significant role of CgEcsit in the C. gigas immune system. Generally, CgEcsit represented a constitutive expression pattern in all the tested tissues, which suggested that CgEcsit might potentially be involved in multiple physiological functions in different tissues of C. gigas.
TLR pathway is of major importance during innate immunity (Akira, Takeda & Kaisho 2001; Irazoqui, Urbach & Ausubel 2010). Most genes in TLR pathway are reported to up-regulated in the stress of bacterial (Regnier, Song, Gao, Goeddel, Cao & Rothe 1997; Escoubas, Briant, Montagnani, Hez, Devaux & Roch 1999). CgEcsit, an essential member of this pathway, was found to be significantly up-regulated after a V. anguilarum challenge as well.
In conclusion, a molluscan Ecsit gene (CgEcsit) was identified from C. gigas for the first time. The CgEcsit mRNA transcript showed a constitutive expression pattern in all the tested tissues and its expression in the haemolymph increased significantly after a V. anguillarum challenge. The study provides basal resources for further study of Ecsit and contributes towards the development of better disease management strategies of oyster farming.
Acknowledgments
We thank Wang Tong for RNA subtraction assistance and Dr Lu Wang for technical assistance and good suggestion. This research was supported by the Major State Basic Research Development Program of China (973 program) (No. 2010CB126401) and a grant from the National Natural Science Foundation of China (No. 40730845).




