A proposal for standardization in forensic bovine DNA typing: allele nomenclature of 16 cattle-specific short tandem repeat loci
Summary
In this study, a proposal is presented for the allele nomenclature of 16 polymorphic short tandem repeat (STR) loci (BM1824, BM2113, ETH10, ETH225, INRA023, SPS115, TGLA122, TGLA126, TGLA227, ETH3, TGLA53, BM1818, CSRM60, CSSM66, HAUT27 and ILSTS006) for bovine genotyping (Bos taurus). The nomenclature is based on sequence data of the polymorphic region(s) of the STR loci as recommended by the DNA commission of the International Society of Forensic Genetics for human DNA typing. To cover commonly and rarely occurring alleles, a selection of animals homozygous for the alleles at these STR loci were analysed and subjected to sequence studies. The alleles of the STR loci consisted either of simple or compound dinucleotide repeat patterns. Only a limited number of alleles with the same fragment size showed different repeat structures. The allele designation described here was based on the number of repeats including all variable regions within the amplified fragment. The set of 16 STR markers should be propagated for the use in all bovine applications including forensic analysis.
Introduction
The capability of PCR amplification of a multiplex of species-specific, polymorphic, short tandem repeat (STR) loci has led to the development of a robust set of 16 bovine STR markers (BM1818, BM1824, BM2113, CSRM60, CSSM66, ETH3, ETH10, ETH225, HAUT27, ILSTS006, INRA023, SPS115, TGLA53, TGLA122, TGLA126 and TGLA227). Although recently a large population study in cattle (Bos taurus) was presented for this robust set of 16 bovine-specific STR markers (Van de Goor L.H.P., Koskinen M.T. & Van Haerignen W.A., unpublished data), no details have yet been published about allele structure or DNA sequence variation in the alleles. This set of 16 markers, which includes the loci recommended by the International Society for Animal Genetics (ISAG) for bovine parentage testing, is included in commonly used commercial kits (Applied Biosystems and Finnzymes Diagnostics) and routinely employed by bovine genotyping providers. For the purpose of more effectively sharing data and for better use in legal casework, a universal nomenclature system should be developed based on the principles of the human repeat-based nomenclature advocated by the International Society of Forensic Genetics (ISFG) recommendations (Gill et al. 1994, 1997; Bär et al. 1997; Schneider et al. 1998). The commonly used current nomenclature system in cattle for parentage verification cases is based on the measured length of PCR amplicons and has evolved in an informal manner from discussions between laboratories during conferences of the ISAG. This allele nomenclature in cattle is based solely on amplicon length and is internationally regulated within ISAG. However, it has major limitations because of the ability to effectively share data generated using different electrophoretic platforms and the fact that allele designation for new STR loci or alleles will not be automatically available. Allele designations based on number of repeats have been advocated for human DNA typing, but it has yet to be universally accepted for animal genetic typing. Although a proposal for allele nomenclature based on number of repeats and an operational comparison with an allelic reference ladder was published for dogs (Canis familiaris) (Eichmann et al. 2004; Hellmann et al. 2006) and partially for cats (Felis catus) (Lipinski et al. 2007), no effort has been made to adapt the same allele nomenclature to a number of widely used important animal species such as cattle, horses and pigs. To develop a repeat-based nomenclature in cattle, a selection of alleles from a dataset containing 22 cattle breeds (N = 9.738) (Van de Goor et al. unpublished data) was sequenced for the 16 polymorphic dinucleotide STR loci. In this study, a proposal is presented for a repeat-based allele nomenclature based on sequenced alleles.
Materials and methods
Selection of loci
The 16 loci (BM1818, BM1824, BM2113, CSRM60, CSSM66, ETH3, ETH10, ETH225, HAUT27, ILSTS006, INRA023, SPS115, TGLA53, TGLA122, TGLA126 and TGLA227) are used globally for routine bovine genotyping for various purposes such as forensic studies, parentage verification and kinship analysis. The markers are commercially available in two panels known as Bovine Genotypes™ Panels 1.1 and 2.1 (Finnzymes Diagnostics).
Samples and DNA extraction
The samples used in this study were collected between January 2005 and February 2007. Over 9000 DNA profiles covering 22 breeds were available, of which 75% were based on hair root samples, 23% originated from blood samples and 2% were based on semen straws and other sample types. A limited number of breeds originated from the Netherlands, i.e. Brandrood Cattle, Dutch Friesian, Groningen Whiteheaded, Dutch Belted, Verbeterd Roodbont and Maas Rijn IJssel, whereas all other breeds exist and are common throughout Europe and North-America. For each of the 16 STR loci, a selection of at least three of the most frequent alleles in the dataset with 22 cattle breeds was sequenced. For the selected alleles, one animal homozygous for the STR allele was sequenced. For the four loci ETH3, ETH225, SPS115 and TGLA122, additional samples or alleles were sequenced as well.
For ETH3, the alleles present in the population data reveal a gap between the three alleles with the smallest observed amplicon lengths. The smallest allele was sequenced to confirm its repeat motif. As one allele of STR ETH225 has been discussed extensively within the ISAG nomenclature, being called either allele 158 or 160, this allele has been selected for sequencing in several animals. In the dataset, only one intermediate allele was observed (STR SPS115) and sequenced. As STR TGLA122 reveals a very extensive size range, the alleles with the largest amplicon lengths have also been selected for sequencing in several animals.
Genomic DNA of the animals was isolated from blood samples or hair roots using routine procedures. For blood samples, 10 μl of blood was washed three times in 150 μl Tris–HCl based buffer. The cell pellet was lysed with proteinase K (0.5 U in 10 mm Tris, 0.5% Tween for 45 min at 56 °C followed by heat inactivation). For hair roots, approximately eight follicles were placed into a PCR tube and lysed with proteinase K (6 U in 10 mm Tris, 0.5% Tween overnight at 56 °C followed by heat inactivation).
PCR and allele sequencing
The 16 STR loci were amplified in a 20-μl volume including 2 μl of DNA (20–150 ng per reaction) using unlabelled primers with the following reagents: 1× GeneAmp PCR Buffer including MgCl2 1.5 mm (Applied Biosystems), 10 pmol of each primer and Taq Gold (Applied Biosystems). The PCR was performed in a 9700 Thermocycler (Applied Biosystems); cycling conditions consisted of 15 min at 95 °C followed by 32 cycles of 30 s at 95 °C, 30 s at 60 °C and 60 s at 72 °C. For the majority of loci, the primer sequences shown in Table 1 were used for the amplification; however, to enable sequencing of the repeats in which the primers were too close to the repeat sequence, new primers were designed for loci where the primers were located within 30 bp of the repeat sequence. For some markers, the primer sequence was partially located in the repeat sequence. Following visualization of an aliquot of the PCR product on an agarose gel, the remaining PCR product was purified with ExoSAP-IT (USB Europe GmbH) treatment according to the manufacturer’s instructions. For each purified PCR product, direct sequencing reactions were performed for the forward and reverse strand using the BigDye terminator v1.1 cycle sequencing kit (Applied Biosystems) according to the manufacturer’s instructions. The sequence reactions were purified using Xterminator (Applied Biosystems) and visualized on an ABI 3100 sequencer. The trace files were analysed using the sequencing analysis 3.7 software (Applied Biosystems) according to the manufacturer’s instructions.
| Locus | Chromosome location | Repeat structure | Repeat sequence | Original references | Primer sequences (Forward and Reverse) | Amplicon length (bp) |
|---|---|---|---|---|---|---|
| BM1818 | D23S21 | Simple | (TG)n | Bishop et al. (1994) | F: AGCTGGGAATATAACCAAAGGR: AGTGCTTTCAAGGTCCATGC | 253–277 |
| BM1824 | D1S34 | Simple | (GT)n | Barendse et al. (1994) | F: GAGCAAGGTGTTTTTCCAATCR: CATTCTCCAACTGCTTCCTTG | 176–188 |
| BM2113 | D2S26 | Simple | (CA)n | Sunden et al. (1993) | F: GCTGCCTTCTACCAAATACCCR: CTTCCTGAGAGAAGCAACACC | 124–146 |
| CSRM60 | D10S5 | Simple | (AC)n | Baylor College of Medicine Human Genome Sequencing Center (2006) | F: AAGATGTGATCCAAGAGAGAGGCAR: AGGACCAGATCGTGAAAGGCATAG | 91–117 |
| CSSM66 | D14S31 | Simple | (AC)n | Barendse et al. (1994) | F: AATTTAATGCACTGAGGAGCTTGGR: ACACAAATCCTTTCTGCCAGCTGA | 177–203 |
| ETH3 | D19S2 | Compound | (GT)nAC(GT)6 | Solinas-Toldo et al. (1993) | 1F: GAACCTGCCTCTCCTGCATTGG1R: ACTCTGCCTGTGGCCAAGTAGG | 100–128 |
| ETH10 | D5S3 | Simple | (AC)n | F: GTTCAGGACTGGCCCTGCTAACAR: CCTCCAGCCCACTTTCTCTTCTC | 206–222 | |
| ETH225 | D9S2 | Compound | (TG)4CG(TG)(CA)n | Steffen et al. (1993) | F: GATCACCTTGCCACTATTTCCTR: ACATGACAGCCAGCTGCTACT | 139–157 |
| HAUT27 | D26S21 | Simple | (AC)n | Thieven et al. (1997) | F: TTTTATGTTCATTTTTTGACTGGR: AACTGCTGAAATCTCCATCTTA | 137–155 |
| ILSTS006 | D7S8 | Simple | (GT)n | Brezinsky et al. (1993) | F: TGTCTGTATTTCTGCTGTGGR: ACACGGAAGCGATCTAAACG | 279–297 |
| INRA023 | D3S10 | Simple | (AC)n | Vaiman et al. (1994) | F: GAGTAGAGCTACAAGATAAACTTCR: TAACTACAGGGTGTTAGATGAACTC | 201–225 |
| SPS115 | D15 | Compound | (CA)nTA(CA)6 | Baylor College of Medicine Human Genome Sequencing Center (2006) | F: AAAGTGACACAACAGCTTCACCAGR: AACCGAGTGTCCTAGTTTGGCTGTG | 247–261 |
| TGLA53 | D16S3 | Compound | (TG)6CG(TG)4(TA)n | Georges & Massey (1992) | F: GCTTTCAGAAATAGTTTGCATTCAR: ATCTTCACATGATATTACAGCAGA | 151–187 |
| TGLA122 | D21S6 | Compound | (AC)n(AT)n | 1F: AATCACATGGCAAATAAGTACATACR: CCCTCCTCCAGGTAAATCAGC | 136–182 | |
| TGLA126 | D20S1 | Simple | (TG)n | 1F: CTAATTTAGAATGAGAGAGGCTTCT R: TTGGTCCTCTATTCTCTGAATATTCC | 111–127 | |
| TGLA227 | D18S1 | Simple | (TG)n | 1F: GGAATTCCAAATCTGTTAATTTGCT1R: ACAGACAGAAACTCAATGAAAGCA | 76–104 |
- 1Primer sequence was re-designed for sequencing; modified primer sequences are not shown.
Nomenclature
For each STR locus, the sequence was compared with the original GenBank sequence. The proposed nomenclature for the 16 bovine STR loci is based on the number of repeat units and the recommendations for human STR nomenclature (Gill et al. 1994, 1997; Bär et al. 1997; Schneider et al. 1998). Table 2 shows an overview of the principles of the human-based nomenclature approach, which has been used for the repeat-based nomenclature applied in this paper. A conversion from the amplicon length-based ISAG nomenclature to the newly presented repeat-based nomenclature is proposed.
| 1. DNA sequences are read in the 5′–3′ direction. |
| 2. For STRs within protein coding genes, the coding strand should be used. For STRs without any connection to protein coding genes, the sequence originally described in the literature or the first public database entry should be used. |
| 3. The repeat sequence motif must be defined so that the first 5′-nucleotides that can define a repeat motif are used. |
| 4. STR repeats are subdivided into three categories based on their repeat structure: |
| Simple repeats: the repeat sequence contains only one motif (e.g. CAn: CA12 = 12). |
| Compound repeats: the repeat sequence varies (e.g. CAnCTn: CA12CT2 = 14). |
| Complex repeats: the repeat sequence contains several different types of repeat motifs, e.g. tetrameric combined with trimeric and/or dimeric motifs (e.g. CAGGnCATnCTn: CAGG2CAT12CT2 = 16). |
| 5. For intermediate alleles, the designation should be based on the number of complete repeats followed by, the number of additional basepairs, each separated by a decimal point. |
Results
Previously, all STR loci included in this study were described as dinucleotide repeat sequences (Table 1 shows locus name, chromosomal location, repeat structure and sequence, original reference, primer sequences and size range of the amplicon length). All STR loci revealed a simple or compound variable repeat structure. The proposed nomenclature for the 16 bovine STR loci investigated is based on the number of repeat units and is adopted from the recommendations of the ISFG for the nomenclature of human STRs.
In the following section (1-3 and S1), the investigated STR loci are presented. They are described by (i) the UniSTS identification number (ii) the GenBank accession number, (iii) the general sequence structure including the flanking regions and (iv) the average allele frequency distribution as observed in the dataset (Van de Goor et al. unpublished data). Sequenced alleles are indicated by an asterisk (*), whereas alleles which were not sequenced have been extrapolated based on the allele mobilities from the raw data. Although deduced allele designation will only be operationally correct, additional sequencing of alleles may not be required for routine analyses as these are based on the application of control samples. If this is confirmed, the STR is described according to previous published studies.
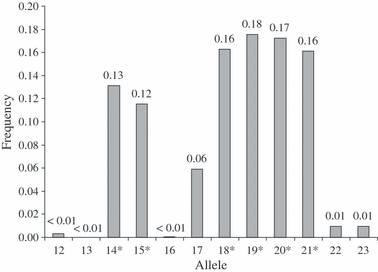
BM2113. Six alleles were sequenced (14, 15, 18, 19, 20 and 21). The sequence corresponds with GenBank accession M97162. The alleles of the locus BM2113 displayed the dinucleotide repeat structure (CA)n. All observed alleles in the sample population clustered into 12 categories; no intermediate alleles were found. Allele 19 was the most frequent with a frequency of 0.18. Accession number for UniSTS: 250697; GenBank: M97162. Sequence: GCTGCCTTCTACCAAATACCCCCTGCTCCGGCCCCCACCTCAAC(CA)n GAGTGAGCTCATAGTCTTGAGTTAAAAAAGTGACAGGTGTTGCTTCTCTCAGGAAG. *, sequenced alleles.
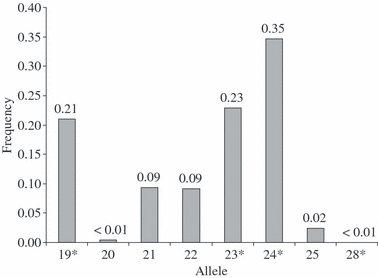
ETH225. Four alleles were sequenced (19, 23, 24 and 28). The sequence corresponds with GenBank accession Z14043. The alleles of the locus ETH225 displayed a compound repeat structure with dinucleotide repeats (TG)4CG(TG)(CA)n. The allele with (TG)4CG(TG)(CA)17 has been designated 23. Furthermore, we observed a single nucleotide polymorphism (SNP) adjacent to the 3′ end of the repeat structure. This C/T polymorphism has no impact on the nomenclature of the locus. The alleles 19, 23 and 24 revealed the T-nucleotide at the SNP position, allele 28 was sequenced in six animals all showing the C-nucleotide at the SNP position. All observed alleles in the sample population clustered into eight categories; no intermediate alleles were found. Allele 24 was the most frequent at 0.35. Accession number for UniSTS: 250852; Genbank: Z14043. Sequence: GATCACCTTGCCACTATTTCCTCCAACATA(TG)4CG(TG)(CA)n[C/T]GATAGCCACTCCTTTCTCTAATGCCACAGAATTACACAGTCAACTCTCTAGTAGCAGCTGGCTGTCATGT. *, sequenced alleles.
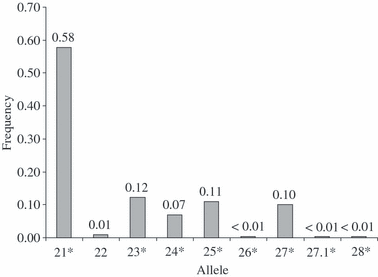
SPS115. Eight alleles have been sequenced (21, 23, 24, 25, 26, 27, 27.1 and 28). All alleles sequenced are consistent with the repeat sequence of GenBank: NW_001503418. The allele structure is that of the compound sequence (CA)nTA(CA)6. According to the guidelines of the International Society of Forensic Genetics, e.g. allele (CA)14TA(CA)6 was designated as allele 21. All observed alleles except one in the sample population clustered into eight categories; the one allele was intermediate between the alleles 27 and 28. This intermediate allele with repeat structure (CA)20TA(CA)6, designated as 27.1, contained one additional A-nucleotide in a stretch of 10 A-nucleotides, 32 nucleotides 3′ upstream of the repeat. Allele 21 was the most frequent, with a frequency of 0.58. Accession number for UniSTS: 279634; GenBank: NW_001503418. Sequence: AAAGTGACACAACAGCTTCACCAGAGCATCTCCAATATCT(CA)nTA(CA)6TCTCATTCCTCTAGTGTCTTTTGCCTTTAAAGAAAAAAAAACTAAGCAGATCAACATGGGATCTCCTTTTTGTAGATTTATAGAAAGGGTTCCTTTGTTGCGCACTCACTTGTAAGAAAATGAGACAAAAACGTGAAACCCACAGCCAAACTAGGACACTCGGTT. *, sequenced alleles.
Discussion
Dimeric STRs are used for bovine genetic identity testing. Because dimeric STRs are more susceptible than trimeric or tetrameric STRs to slippage during PCR amplification resulting in the generation of stutter bands, the human forensic community chose larger repeat motif STRs, predominately tetrameric STRs. In contrast, for parentage verifications and identity testing in bovines, historically dimeric STRs have been selected because tetranucleotide loci were not available initially. There is less need to change tousing tetrametric STRs in cattle, because of (i) the extensive, global databases with cattle genotypes based on dimeric STRs; (ii) the optimization of amplification conditions that reduce stutter production for these loci; (iii) efforts to implement Single Nucleotide Polymorphisms in cattle genotyping; and (iv) the use of the Phusion DNA-polymerase in the bovine STR panels provides proofreading activity that reduces the tendency observed with DNA-polymerases for non-template adenylation of PCR products (+A). Therefore, the current bovine STR loci are likely to be used in the foreseeable future for identity testing.
The preferred method for naming STR alleles in the human DNA forensic science community is based on the number of repeats (Gill et al. 1994, 1997; Bär et al. 1997; Schneider et al. 1998) and this is applied operationally by comparison with an allelic ladder. Previously, the motif structure had not been described for the dinucleotide STR loci used in bovine genetic identity testing. This is necessary to develop a consistent nomenclature and for generating a community-wide allelic ladder. The repeat structure of the bovine STRs has been described herein by sequence analysis of a sampling of the alleles at each locus.
Homozygous profiles of alleles at each STR locus were directly sequenced from the purified PCR product; no alleles from heterozygote profiles were used. From the 16 investigated loci, 11 showed a simple repeat structure, whereas compound repeat structures were present in the five loci ETH3, ETH225, SPS115, TGLA53 and TGLA122 (2, 3 & S1). None of the 16 STRs showed complex repeat sequences. There were a few inconsistencies in sequence with those in GenBank, but these were predominately in the flanking region and did not affect the repeat structure. The inconsistencies are primarily caused by the original sequence methods, which were used approximately 15 years ago. Allele designation was straightforward in almost all cases, as only one intermediate allele (a single nucleotide variant –SPS115 allele 27.1) was observed in our dataset of 9738 individuals. Considering the low number of intermediate alleles found in the large dataset (>9000 animals covering 22 breeds), the number of other intermediate alleles not yet identified should remain low.
In Table 3, a conversion is provided for the amplicon length-based allele nomenclature in cattle, which is internationally recommended within ISAG. One allele of STR ETH225 has been extensively discussed within the ISAG nomenclature, either being called allele 158 or 160 depending on the analytical conditions used. This allele was identified by our sequencing results to have a repeat-based nomenclature of 28.
| Repeat number | ISAG | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BM1818 | BM1824 | BM2113 | CSRM60 | CSSM66 | ETH3 | ETH10 | ETH225 | HAUT27 | ILSTS006 | INRA023 | SPS115 | TGLA53 | TGLA122 | TGLA126 | TGLA227 | |
| 11 | 75 | |||||||||||||||
| 12 | 178 | 121 | 77 | |||||||||||||
| 13 | 256 | 180 | 123 | 198 | 79 | |||||||||||
| 14 | 258 | 182 | 125 | 88 | 209 | 140 | 200 | 137 | 109 | 81 | ||||||
| 15 | 260 | 184 | 127 | 90 | 179 | 103 | 142 | 202 | 139 | 111 | 83 | |||||
| 16 | 262 | 129 | 92 | 181 | 213 | 144 | 284 | 204 | 141 | 113 | 85 | |||||
| 17 | 264 | 188 | 131 | 94 | 183 | 215 | 146 | 286 | 206 | 143 | 115 | 87 | ||||
| 18 | 266 | 190 | 133 | 96 | 185 | 109 | 217 | 148 | 288 | 208 | 145 | 117 | 89 | |||
| 19 | 268 | 135 | 98 | 187 | 219 | 140 | 150 | 290 | 210 | 154 | 147 | 119 | 91 | |||
| 20 | 270 | 137 | 100 | 189 | 221 | 142 | 152 | 292 | 212 | 156 | 149 | 121 | 93 | |||
| 21 | 272 | 139 | 102 | 191 | 115 | 223 | 144 | 154 | 294 | 214 | 248 | 158 | 151 | 123 | 95 | |
| 22 | 141 | 104 | 193 | 117 | 225 | 146 | 156 | 296 | 216 | 250 | 160 | 153 | 125 | 97 | ||
| 23 | 143 | 106 | 195 | 119 | 148 | 158 | 298 | 218 | 252 | 162 | 155 | 99 | ||||
| 24 | 108 | 197 | 121 | 150 | 300 | 220 | 254 | 164 | 157 | 101 | ||||||
| 25 | 280 | 199 | 123 | 152 | 302 | 222 | 256 | 166 | 159 | 103 | ||||||
| 26 | 201 | 125 | 258 | 168 | 161 | |||||||||||
| 27 | 114 | 127 | 260 | 170 | 163 | |||||||||||
| 27.1 | 261 | |||||||||||||||
| 28 | 205 | 129 | * | 262 | 172 | 165 | ||||||||||
| 29 | 131 | 174 | 167 | |||||||||||||
| 30 | 176 | 169 | ||||||||||||||
| 31 | 178 | 171 | ||||||||||||||
| 32 | 180 | 173 | ||||||||||||||
| 33 | 182 | 175 | ||||||||||||||
| 34 | 184 | 177 | ||||||||||||||
| 35 | 186 | 179 | ||||||||||||||
| 36 | 188 | 181 | ||||||||||||||
| 37 | 190 | 183 | ||||||||||||||
- ISAG, International Society of Animal Genetics.
- *No agreement within ISAG about nomenclature of this allele, allele is being called 158 or 160 depending on electrophoretic conditions used.
In the human forensic community, the recommendations by ISFG (Gill et al. 1994, 1997; Bär et al. 1997; Schneider et al. 1998) include the use of standardized allelic ladders to enable designation of alleles in samples. At present, unfortunately no allelic ladders for the 16 STRs are commercially available. Thus, laboratories performing STR typing will have to base their allele calls only on an internal size standard and one control sample. However, the data from these allele sequences will be useful for future efforts to develop an allelic ladder for all 16 loci.




