RUNX1: transcription factor scores a hat-trick of autoimmune diseases
An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis
Tokuhiro et al. (2003)
Nature Genetics 35: 341–348
A putative RUNX1 binding site variant between SLC9A3R1 and NAT9 is associated with susceptibility to psoriasis
Helms et al. (2003)
Nature Genetics 35: 349–356
A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans
Prokunina et al. (2002)
Nature Genetics 32: 666–669
For a long time, researchers have speculated that autoimmune diseases may share common genetic factors. Now, the evidence that supports this hypothesis has surfaced. Less than a year ago, Prokunina et al. reported that a single nucleotide polymorphism (SNP) in the gene PDCD1 is associated with an increased risk of systemic lupus erythematosus (SLE). The authors show that this SNP leads to an alteration in a binding site for the runt-related transcription factor 1 (RUNX1, also known as AML1). Inspired by these findings, researchers have begun examining the role of RUNX1 in other autoimmune diseases. These efforts have lead to the discovery that mutations in RUNX1-binding sites in two additional genes are associated with two other autoimmune diseases, psoriasis and rheumatoid arthritis (RA).
Although SLE, psoriasis, and RA are all part of the family of autoimmune diseases, they have very different clinical presentations. RA (OMIM 180300) is mainly characterized by a chronic inflammation of the (small) joints. Eventually, this leads to destruction of both cartilage and bone, often leaving the patient both deformed and disabled. Psoriasis (OMIM 177900) is predominantly a skin disease. White, silvery plaques that are typical of this disease can cover any part of the body, although usually the scalp, extensor surfaces of elbows and knees and the lower portion of the back are affected. SLE (OMIM 152700) is a multiorgan disease with a very diverse clinical presentation. The most common symptoms are fever, joint pain, rash, and signs of renal disease.
RUNX1 is one of three members of the highly conserved family of runt-domain transcription factors. Together with CBFβ, they form heterodimers that bind to the DNA sequence TGT/CGGT. RUNX1 is highly expressed in hematopoietic cells and is believed to play an important role in the formation and differentiation of these cells. This function is supported by the severe defect in hematopoiesis observed in RUNX1-null mice. In humans, disruption of the RUNX1 gene (by chromosomal translocation) causes acute myeloid leukemia.
The search for the role of RUNX1 in autoimmune diseases was initiated by the work of Prokunina et al. They screened over 2500 SLE patients for SNPs in PDCD1, a gene that has previously been associated with SLE. One of the SNPs, located in intron 4, showed a strong correlation with SLE. Further research revealed that this SNP disrupts a predicted DNA-binding site for RUNX1. Based on their findings and the fact that absence of PDCD1 in mice leads to a phenotype similar to SLE, the authors hypothesized that RUNX1 modulates the expression of PDCD1 by increasing its transcription. Disruption of the RUNX1-binding site would lead to a decreased expression of PDCD1 and thereby contribute to the development of SLE.
In their study, Helms et al. used a haplotype block-based association analysis of psoriasis at the PSOR2 locus on chromosome 17q25. Using a total of 123 markers, they identified two regions that were associated with the disease. In the current report, they describe their findings in the proximal region. This region is located between two genes: SLC9A3R1, a member of the solute carrier family 9, and NAT9, a gene with an unknown function. The two genes lie in a tail-to-tail orientation and are separated by only 1.2 kb. Five SNPs responsible for the association between the overtransmitted haplotypes and psoriasis were then examined further in detail. Because all SNPs were located in intronic DNA, the authors examined whether any of the SNPs caused a change in a transcription factor-binding site. This analysis revealed that the psoriasis-associated allele of one of the SNPs eliminated a binding site of RUNX1. Binding assays showed that indeed the wildtype allele binds RUNX1, and this binding is abolished in the psoriasis allele. Despite various experimental approaches, the authors were unable to discover which of the two genes is affected by the mutation, and therefore, further studies are required to determine how RUNX1 is involved in the development of psoriasis.
Tukihiro et al. applied a similar approach in their study. By using case-control linkage disequilibrium mapping on a total of 115 SNPs, they identified a SNP in the gene SLC22A4 that was associated with an increased risk of RA. As with the other two studies, this SNP also caused a change in an RUNX1-binding site. Tukihiro et al. then went one step further. They hypothesized that if a change in an RUNX1-binding site affects RA, it is also likely that expression differences of RUNX1 itself would correlate with RA. Analysis of correlation of SNPs in the RUNX1 gene with RA proved their hypothesis to be correct. They found a SNP located in intron 6 that was significantly associated with RA, thus providing additional evidence of a direct role of RUNX1 in RA.
In summary, these publications provide a new mechanism that connects three different autoimmune diseases in which RUNX1 plays a leading part (Fig. 3). RUNX1 and CBFβ, by forming a heterodimer, form the transcription factor, core binding factor (CBF). CBF can act both as an enhancer or a repressor by binding to the specific DNA sequence TGT/CGGT. So far, three different autoimmune diseases have shown to be associated with mutations in RUNX1-binding sites. However, the search has only just begun and it seems likely that other diseases will soon follow.
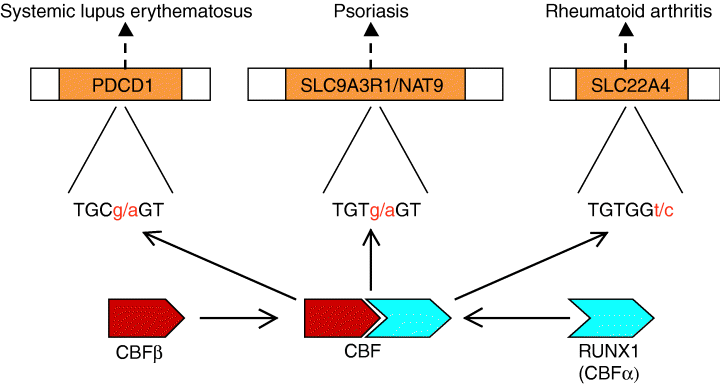
Schematic example of the role of RUNX1 in autoimmune diseases.




