The quality of organic matter shapes the functional biogeography of bacterioplankton across boreal freshwater ecosystems
Abstract
Aim
The need to go beyond taxonomy to understand patterns in microbial function has led to an increased use of trait-based approaches, yet we know little about how microbial functional traits vary across large-scale environmental gradients in natural ecosystems. Here, we apply a trait-based approach to explore the large-scale variability in the trait structure underlying the processing of dissolved organic matter (DOM) by boreal bacterioplankton communities, as well as its regulation and links to taxonomic composition.
Location
Samples were collected from 296 rivers and lakes across five regions in northern Quebec (Canada), which span large gradients in environmental, climatic and geographical properties typical of the boreal zone.
Methods
We used the metabolic profiles obtained with Biolog EcoPlates® as an imprint of the trait structure underlying bacterial processing of DOM, and Illumina sequencing of the 16SrRNA gene to characterize the taxonomic composition of these bacterial assemblages. The resulting spatial patterns were compared with an array of climatic, landscape and limnological properties varying at the landscape scale.
Results
Despite a clear regional segregation of the sampled sites based on environmental variables, the trait structure of boreal bacteria did not show any regional or ecosystem-specific patterns, but rather was linked to a gradient of quality of DOM. Community trait configurations diverged progressively with decreasing terrestrial influence, probably due to local processes that transform and diversify the available pool of DOM. This DOM quality gradient did not explain the taxonomic biogeography of these communities, which was controlled by a different set of environmental factors.
Main conclusions
The functional biogeography of boreal bacterioplankton is driven by the nature of the DOM pool, and particularly by the influence of terrestrial DOM. The lack of coherence between functional and taxonomic biogeographies implies that the environmental controls of freshwater bacterial performance cannot be directly inferred from spatial patterns in taxonomic composition.
Introduction
Microbes play fundamental roles in ecosystems by mediating biochemical cycles, and much effort has been devoted to understanding how microbial communities are assembled in nature and carry out processes of ecological significance. Studies investigating the relationship between the taxonomic composition and ecosystem functioning, however, have yielded inconclusive results (e.g. Langenheder et al., 2005; Lear et al., 2014), suggesting that taxonomic composition may not always provide insight into microbial processes at the ecosystem level. Regardless of the reasons for this apparent uncoupling between composition and function (e.g. dormancy, lateral gene transfer, functional redundancy; Ochman et al., 2000; Allison & Martiny, 2008; Jones & Lennon, 2010), there is a clear need to go beyond a taxonomic characterization of communities and towards an exploration of the functional properties that underlie microbial performance in natural ecosystems.
It has recently been emphasized that trait-based approaches, which investigate variations in functional attributes related to community performance, might eventually bridge the gap between microbial taxonomy and ecosystem functioning (Green et al., 2008; Wallenstein & Hall, 2012; Krause et al., 2014; Nemergut et al., 2014). However, because of the difficulties of transferring the principles of trait-based approaches to complex natural microbial assemblages, the application of trait-based approaches to the microbial world remains largely theoretical, restricted to small scales or purely experimental (see references in Krause et al., 2014). This limits our ability to use such functional attributes for understanding patterns in major microbially mediated ecosystem processes.
Traditionally, trait-based functional ecology has relied on the selection of traits that are relevant to community performance, assigning them to extant taxa to derive aggregate weighted community values for each trait, and exploring their intra- or intercommunity variability (Swenson & Enquist, 2007; Vogt et al., 2013; Violle et al., 2014). In the case of complex microbial assemblages, however, it is virtually impossible to assign traits to the thousands of taxa typically detected from sequencing of environmental DNA. Therefore, the trait values in these microbial communities cannot be reconstructed from the bottom up (i.e. measured at the individual level; Violle et al., 2007) but need to be determined from the top down, as aggregated functional properties (Wallenstein & Hall, 2012; Fierer et al., 2014; Krause et al., 2014). The challenge for the application of functional trait approaches to microbial communities is thus to identify measurable aggregate properties that can be associated to community traits underlying key bacterial processes. In addition, for these microbial properties to be eventually useful for predictive purposes (e.g. Wallenstein & Hall, 2012), they should be responsive to measurable environmental factors.
Current research is increasingly focusing on the inference of community traits from genomic information (i.e. functional genes and proteins, metabolic versatility, genome size; Green et al., 2008; Burke et al., 2011; Barberán et al., 2012, 2014; Fierer et al., 2014). Although these molecular approaches provide valuable insight into the potential functional diversity of bacterial communities, they are still far from being easily translated into actual bacterial processes. Given the lack of large-scale empirical studies, moreover, we still know little about how the expression of microbial community traits varies over large environmental gradients – a necessary step if we are ever to understand the underlying basis of the functional responses of bacterial communities to environmental forcing. In this paper we address some of these shortcomings by developing a trait-based conceptual framework that allows us to explore the large-scale biogeography and regulation of the functional trait structure linked to a key ecosystem process, the bacterial consumption of dissolved organic carbon (DOC), across boreal aquatic ecosystems. We further assess the potential links between the functional and taxonomic biogeographical patterns of these bacterioplankton communities.
Conceptual framework
Of all bacterial functions, the utilization of DOC is among those with the greatest ecosystem implications because it influences the nature, persistence and fate of carbon in the biosphere (Battin et al., 2008). The bacterial interaction with DOC requires a combination of functional abilities, in this case linked to the uptake and processing of the diverse carbon compounds that make up the complex aquatic DOC pools. Each of these abilities or traits (e.g. the capacity to transport and utilize a certain substrate) will be associated with a specific gene or combination of genes (e.g. genes for transporters and enzymes; Fig. 1a-1), and, depending on the degree of phylogenetic conservatism of those genes (Martiny et al., 2013), these capacities will be more or less widely distributed among the taxa within a community. In turn, the different bacteria carrying the trait in question may have different levels of ability for that trait (e.g. varying uptake capacity for a given substrate), resulting in a distribution of the trait values (sensu Wallenstein & Hall, 2012) among community members (Fig. 1a-2). The ensemble of distributions of trait values associated with a given aggregate community function is what we term ‘community trait structure’ (Fig. 1a-3). Together, the realized processes enabled by the various traits shape the ‘community functional structure’, which in this case comprises the actual uptake kinetics and processing rates for the diverse substrates (Fig. 1a-4). Hence, while the trait structure establishes the potential of the community for carbon acquisition, the functional structure is what determines the realized aggregate ecosystem process (i.e. the actual amount and nature of the carbon processed by the bacterial community; Fig. 1a-5).
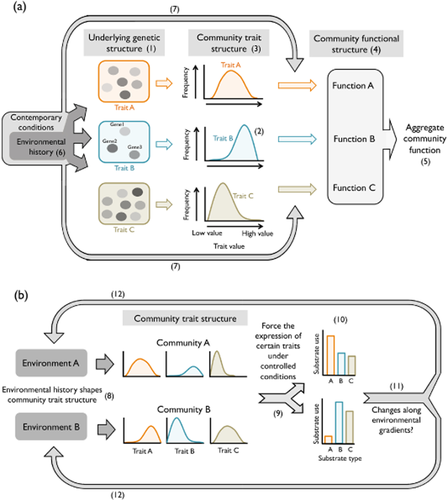
(a) Conceptual scheme of the trait structure underlying major bacterial processes and its environmental regulation. Major bacterial functions (e.g. bacterial utilization of dissolved organic carbon, DOC) result from the combination of multiple abilities (i.e. the ability to use the diverse carbon substrates). Each of these abilities or traits is codified by a combination of genes (1) that can be more or less widely distributed among the pool of taxa. In turn, the different species may have different levels of capacity for each trait (e.g. high or low uptake capacity for a given substrate, 2), resulting in different distributions of the trait values among community members, which collectively make up the ‘community trait structure’ (3). The proportion of cells that are expressing those traits at a given moment will determine the realized function enabled by each trait, which together make up the ‘community functional structure’ (4) and collectively determine the aggregate ecosystem level process (e.g. the total bacterial carbon processing, 5). In this context, the environment affects bacterial performance via two major pathways: historically, by shaping the trait structure through environmental sorting of species (6), and currently, by modulating the expression of these traits through providing substrates or influencing the physiology of the cells (7). (b) The approach of this study is based on exposing bacterial communities that have experienced widely different environmental histories to the same current environmental conditions, in this case, a fixed set of organic carbon substrates under controlled conditions (9). Differences in the resulting patterns of substrate utilization will reflect differences in the original trait configurations of the communities (10). The exploration of the variability in these patterns of DOC use in relation to broad environmental gradients (11) allows us to infer the factors that shaped the underlying trait structure of these bacterioplankton assemblages, providing further insight into the scales at which this environmental filtering of functional capacities operates at the landscape level (12).
In this context, the environment has a double role in influencing bacterial function. First, it shapes the community trait structure through the sorting of species, such that the trait structure reflects the environmental history of the community, i.e. the conditions to which it has been exposed for a certain period of time (Fig. 1a-6). Second, current environmental conditions may modulate the expression of the trait structure through variations in resource supply and by influencing the physiology of the cells (Fig. 1a-7).
Although in reality we cannot determine the actual distribution of traits within complex bacterial assemblages, this scheme suggests that it may be possible to indirectly assess potential differences in trait structure among communities by assessing their functional response under comparable current environmental conditions, which is the basis of our approach. We conceptualize this approach as it pertains to substrate acquisition in Fig. 1 (b): bacterial communities differing in their environmental history will be characterized by distinct trait structures (Fig. 1b-8) so that, when subjected to the same current conditions (Fig. 1b-9), variations in the functional response will reflect differences in their underlying trait structures. Here we use the differences in the substrate utilization profiles generated by Biolog EcoPlates® (Hayward, CA, USA) which expose bacteria to a fixed set of organic compounds; Fig. 1b-10) as a reflection of the variations in the community trait structure underlying the processing of DOC. We have applied this approach to explore the large-scale spatial patterns in the trait structure underlying the processing of organic carbon by boreal bacterioplankton communities, since the exploration of the variation of these metabolic profiles along large environmental gradients (Fig. 1b-11) allows us to identify the environmental factors that shape the functional trait structure of communities at the landscape scale (Fig. 1b-12). Specifically, we measured these metabolic profiles in 296 lakes and rivers across five boreal regions of Québec (Canada), which span wide environmental, geographic and climatic gradients. Since the key environmental drivers may vary at different spatial scales, our aim was to assess whether bacterial trait structure is discretely structured in nature (as a result of regional or ecosystem-specific environmental histories) or whether it changes along environmental gradients that may transcend regions or ecosystem types. Finally, we compared the observed patterns in trait structure with those of the taxonomic composition of the same communities assessed through 16S rRNA gene sequencing.
Methods
Study sites and sampling design
Samples were collected between 2009 and 2013 from 296 boreal rivers and lakes across five regions in northern Quebec (Canada) spanning large gradients in environmental features and climate (see Appendix S1 in Supporting Information for an overview) and cover a total area of c. 900,000 km2 (44–56° N, 64–80° W; Fig. 2a). Further details on the regional features of the sampled sites are given in Rasilo et al. (2015).
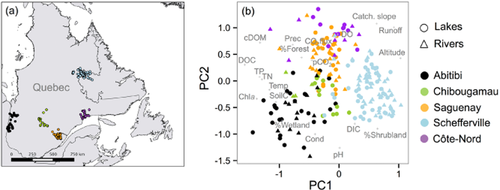
(a) Distribution of sampling sites across the five sampled regions in northern Quebec (Canada). (b) Principal component analysis (PCA) of samples based on all the measured environmental and geographical parameters. Colours/shades indicate the different regions, and shapes the rivers (triangles) or lakes (circles). The two first axes explain 43.5% of the variance. Note that the sampled sites showed a clear regional imprint based on these parameters. DOC, dissolved organic carbon; DIC, dissolved inorganic carbon; cDOM, coloured dissolved organic matter; TP;TN, total phosphorus and nitrogen; Chla, chlorophyll a; Prec, mean annual precipitation; Runoff, mean annual runoff; SoilC, mean soil organic carbon content in the catchment; Catch. Slope, average catchment slope; Altitude, mean catchment altitude; %Forest/%Shrubland/%Wetland, percentage of the area in the catchment covered by forest/shrubland/wetland; Temp, temperature; Cond, conductivity.
All sites were sampled once during summer between June and August and cover a wide range in lake area (0.002–4345 km2) and river order (Strahler order 1–8). Samples were taken at a depth of 0.5 m at the deepest measured spot of lakes and near the shore in rivers. Temperature, dissolved oxygen, pH and conductivity were measured in situ with a YSI probe. Samples were filtered in situ through 0.45 μm PES cartridge filters and stored in acid-washed glass vials for DOC and optical analyses, or kept in the dark in acid-rinsed bottles for further processing.
Biological, chemical and optical analyses
Chlorophyll a (Chla) concentration was determined spectrophotochemically in hot ethanol extracts (90%). Samples for the determination of bacterial abundances were preserved with 1% glutaraldehyde and analysed with a FACSCalibur flow cytometer. The DOC concentration was measured on an OI 1010 TOC analyser. Total phosphorus (TP) and nitrogen (TN) concentrations were analysed after persulphate and alkaline persulphate digestions, respectively, following standard methods (see Rasilo et al., 2015). The surface water ambient concentration of CO2 (pCO2) and air/water CO2 fluxes were measured as in Rasilo et al. (2015). All these measurements were performed in duplicate.
The optical properties of DOC were measured as indices of its composition and source. Coloured dissolved organic matter (cDOM) was quantified as the absorbance at 440 nm using an Ultrospec 3100 spectrophotometer. DOC composition was described on the basis of fluorescence absorption/emission spectra (EEMS) measured in a Shimadzu RF5301 PC spectrofluorophotometer across excitation/emission wavelengths of 275–450 nm and 280–600 nm, respectively. Six main fluorescence components were recovered from the EEMS using parallel factor analysis (PARAFAC): Components 1 to 3 (C1–C3), previously related to refractory, humic material of presumably terrestrial origin, and C4–C6, associated with more biolabile freshly produced DOM (for details see Lapierre & del Giorgio, 2014; Stubbins et al., 2014). In particular, C5 and C6 have been linked to autochthonous processes, C6 having a fluorescence signature typical of protein-like material. The percentage contribution of each component was calculated relative to the total fluorescence of the six PARAFAC components.
Geographical analyses
The lake surface areas, river length and order, catchment areas and the elevation of the sampled sites were derived using the ArcMap 10 and ArcGIS V10 software (ESRI Inc., Redland, CA, USA) applied on the DEM derived from 1:50,000 maps. For each catchment area, we averaged the slope based on the DEM, as well as various land-cover properties obtained from Geobase (2009). Mean annual temperature, precipitation and runoff were extracted at each site location from a long-term climate database (WordClim; Hijmans et al., 2005). The water residence time of lakes was estimated from lake volume (estimated as mean depth × lake area), catchment area and mean annual regional runoff. River residence time was calculated based on the measured water velocity at the sampling point and the total upstream distance.
Bacterial substrate utilization patterns
We used the substrate utilization patterns obtained with Biolog EcoPlates® as a simplified expression of the underlying trait structure associated with utilization of DOC by bacteria. Unfiltered water (125 μl) from each site was added to each well of the EcoPlates®, which contain 31 different carbon sources in triplicate. The bacterial respiratory activity associated with substrate use reduces a tetrazolium dye, producing colour measurable as the absorbance at 595 nm. One plate per site was incubated in the dark and at room temperature for 2 to 9 days depending on colour development rates. The absorbance was measured periodically until an asymptote was reached. At each time point, the overall colour development of the plates was expressed as the average well colour development (AWCD), and the mean colour development of each compound was calculated as the blank-corrected mean absorbance of each substrate measured at the time when the AWCD was closest to 0.5 (Garland et al., 2001), usually between days 1 and 4. We also registered the maximum AWCD attained by each sample as a proxy of the maximum average substrate utilization.
Bacterial community composition
At 274 of the 296 sites, 300–500 ml samples were filtered onto 0.22 μm pore-size filters. Genomic DNA was extracted using the MoBio PowerWater DNA extraction kit following the manufacturer's protocol. The V3–V4 region of the 16SrRNA gene was amplified with the primers 515F and 806R, and sequenced on an Illumina MiSeq2000 following a paired-end approach (Caporaso et al., 2012). Paired-end reads were assembled with flash (Magoc & Salzberg, 2011) and sequences between 250 and 290 bp were used for downstream analyses in qiime in order to remove primers, low-quality, archaeal and chloroplast reads, and singletons (Caporaso et al., 2010). Quality sequences were binned into operational taxonomic units (OTUs; ≥ 97% similarity) using uclust v.1.22q (Edgar, 2010), and representative sequences were classified using the Ribosomal Database Project taxonomy (Wang et al., 2007). To enable comparisons between samples, the OTU table was randomly subsampled to ensure an equal number of sequences per sample, based on the sample with the fewest reads (8415 sequences).
Statistical analyses
To detect significantly different groups based on the functional properties of communities, we performed k-means clustering using the Hartigan–Wong algorithm (Hartigan & Wong, 1979). The Bray–Curtis distance was used as an estimator of functional or taxonomic dissimilarity, and was visualized by non-metric multidimensional scaling (NMDS) analysis. Differences in environmental, functional or taxonomic structure between categories (i.e. regions, ecosystems) were tested with anosim (analysis of similarity, Clarke, 1993), performing 10,000 permutations based on either Euclidean (environmental data) or Bray–Curtis (functional and taxonomic data) distances. The ‘envfit’ function (R Vegan package) was used to explore the correlations between different environmental variables and functional or taxonomic ordination patterns. The significance of the associations was determined by 999 random permutations. All analyses were performed in JMP 9.0.1 (SAS Institute, NC, USA) or R 3.0.0 software (R Core Team, 2013).
Results
Distribution of bacterial trait structure across large spatial gradients
The aquatic ecosystems studied here encompass large variations in climate, landscape and limnological properties (Appendix S1, Fig. 2a,b). The distribution of sites based on the main measured environmental and landscape properties (Fig. 2b) showed a clear segregation among the studied regions (anosimBY REGION, R = 0.61, P < 0.0001). In general, rivers and lakes within one region were more similar to each other environmentally than systems from different regions. Briefly, regions differed mostly in terms of DOC concentration and trophic status (captured by PC1), ranging from the most productive waters in the Abitibi region, to the most oligotrophic sites in Schefferville. In addition, sites aligned along a gradient of catchment slope and mean annual runoff (PC2), which also corresponded to changes in conductivity and pH. Despite these clear regional differences in environmental and topographic parameters, we found no comparable spatial patterns in community trait structure based on substrate uptake profiles (Fig. 3a). In contrast, lake and river bacterial trait structures seemed to segregate to a certain extent (anosimBY ECOSYSTEM, R = 0.14, P < 0.001; Fig. 3a) regardless of the geographical location, although there was a large degree of overlap between both types of ecosystem.
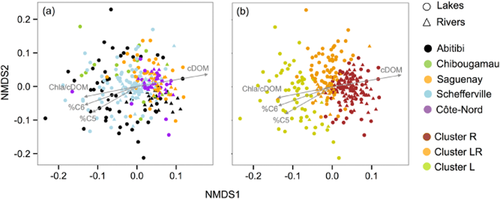
Non-metric multidimensional scaling (NMDS) ordination of samples based on the trait structure of bacterioplankton communities. In both cases, triangles indicate rivers and circles, lakes. Samples are labelled according to geographical region (a) or functional cluster (i.e. k-means classification at three groups partition) (b). The environmental variables showing the strongest correlations (R = −0.36–0.53, P < 0.001) were fitted to the ordination. cDOM, coloured dissolved organic matter; Chla/cDOM, ratio of chlorophyll a concentration to cDOM; %C5,%C6, percentage contribution of the fluorescent parallel factor analysis components C5 and C6 to the total fluorescence.
Drivers of bacterial trait structure over large landscape gradients
To determine the factors linked to the observed patterns in trait structure, we performed linear regression analysis between the NMDS scores and all the measured environmental parameters. Variables associated to DOM concentration and quality emerged as the most strongly related to the ordination (Fig. 3a). The concentration of cDOM, indicative of terrestrial influence (Lapierre & del Giorgio, 2014), the relative proportions of fluorescent components C5 and C6 (associated with freshly produced labile DOM) and the ratios of Chla to cDOM (a proxy of the relative proportions of terrestrial and autochthonous inputs; Carpenter et al., 2005) were the variables with strongest (but opposite) correlations with both NMDS axes (Fig. 3a).
These results, together with the large degree of functional overlap between lake and river communities, suggest that rather than being ecosystem specific, the trait structure of bacterial communities varies along the continuum of DOM quality and quantity represented by NMDS axis 1 (Fig. 3a). A k-means classification algorithm supported this observation by separating the sites into three functionally distinct groups that also aligned along axis 1 (Fig. 3b). The resulting clusters comprised either mostly rivers (cluster R; 71% rivers, 29% lakes), a mixture of lakes and rivers (cluster LR; 68% lakes, 32% rivers) or mostly lakes (cluster L; 95% lakes, 5% rivers). Besides changes in the relative use of the 31 Biolog substrates (details not shown), these three clusters of communities also differed in the number of substrates used, which was slightly higher in clusters R and LR than in L sites (Table 1). Moreover, while the distribution of substrate use was more even in L systems, the maximum average substrate use (maximum AWCD) increased markedly towards R systems (Table 1).
| Cluster R | Cluster LR | Cluster L | |
|---|---|---|---|
| Biolog properties | |||
| Number of substrates used | 29.6 (0.2)a | 29.6 (0.2)a | 28.2 (0.3)b |
| Functional evenness (Pielou's) | 0.950 (0.001)a | 0.956 (0.001)b | 0.962 (0.002)c |
| Maximum average substrate use | 1.65 (0.03)a | 1.29 (0.03)b | 0.94 (0.04)c |
| Measured site characteristics | |||
| cDOM (a440; m−1) | 5.0 (0.3)a | 2.1 (0.4)b | 1.5 (0.5)c |
| cDOM/DOC | 0.5 (0.02)a | 0.3 (0.02)b | 0.2 (0.03)c |
| Chla/cDOM | 0.7 (0.2)a | 1.0 (0.2)b | 2.5 (0.2)c |
| %C6 | 10.3 (0.7)a | 14.8 (0.9)b | 19.5 (1.1)c |
| Catchment area (km2) | 84.2 (22.2)a | 5.9 (3.5)b | 0.07 (0.07)c |
| Catchment slope (º) | 5.9 (0.3)a | 4.5 (0.3)a | 3.6 (0.4)b |
| Water residence time (days) | 169 (47)a | 370 (86)b | 975 (268)c |
- cDOM, coloured dissolved organic matter; DOC, dissolved organic carbon; Chla, cholrophyll a; ,%C6, percentage contribution of the fluorescent parallel factor analysis component C6 to the total fluorescence.
A comparison of the variables characterizing the sites within each cluster further supports the hypothesis that the biogeography of community trait structure is linked to a gradient of terrestrial influence on the DOM pool: sites belonging to the R cluster were characterized by significantly higher concentrations of cDOM and higher ratios of cDOM to DOC compared with L systems (Table 1). These sites had significantly larger and steeper catchments, indicating a stronger connectivity to land and shorter residence times than for L sites (Table 1). Conversely, sites from cluster L showed much higher ratios of Chla to cDOM and greater proportions of C6 than R sites, suggesting an increase in the relative importance of autochthonous sources of DOM in theses sites, and a weaker connection to land. There were no consistent differences between clusters in any of the other measured limnological and geographical variables.
Diversification of the trait structure along the DOM gradient
The ordination of sites based on community trait structure as reflected by the Biolog profiles (Fig. 3) further shows that while lake communities were distributed across the entire ordination space, river assemblages clustered more closely. In order to explore whether the degree of dispersion varied in relation to the gradients in DOM, we ranked our sites according to their respective Chla to cDOM ratio, binned the data into 10 equal groups of 26 or 27 sites and calculated the mean Bray–Curtis distance (in terms of their metabolic profiles) between communities within each bin. Higher values of this distance indicate greater functional differences between communities within a given bin. This exercise clearly shows that functional dissimilarity was highest within bins that had the least terrestrial influence, and declined steeply with increasing terrigenous inputs, evidenced by both a strong negative relationship with cDOM (Fig. 4a) and by a positive relationship with the C6 component of DOM (Fig. 4b). In addition, the functional dissimilarity between communities was also higher within bins characterized by a more diverse DOM pool, as reflected by the Shannon index based on the six fluorescent components of DOM (Fig. 4c).
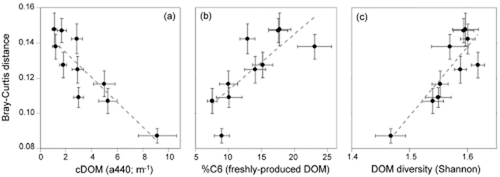
Relationships between the dissimilarity (Bray–Curtis distance) in the trait structure of communities and averaged values of coloured dissolved organic matter (cDOM) (a), percentage contribution of the fluorescent parallel factor analysis component C6 to the total fluorescence (%C6) (b) and diversity of DOM (calculated as the Shannon index of the six fluorescent components) (c). The 266 sites were ordered based on their chlorophyll a/cDOM ratio and binned into 10 equal groups (n = 26–27), for which means and standard errors of either the distance or the environmental parameters were calculated. Note that the functional dissimilarity between sites is higher in systems with lower cDOM, and higher %C6 and DOM diversity. R2 = 0.77, 0.59, and 0.74, respectively, P < 0.0001.
Comparison between patterns in functional trait structure and bacterial community composition
Overall we obtained 2,305,710 good quality sequences, which clustered into 155,578 OTUs. The description of the spatial patterns in bacterial community composition is the focus of a companion paper in preparation by J.P.N.-G. and colleagues, but in brief, an NMDS based on Bray–Curtis distances showed that whereas there was some regional structuring, rivers and lakes showed a much clearer segregation and much less overlap between them (Appendix S2) than found on the basis of the functional patterns. As a result, the functional and taxonomic distance matrices were not significantly correlated (P > 0.05, Mantel test). We further compared the ordination patterns of the functional and taxonomic data by relating the respective NMDS axes (Fig. 5a,b), and found that the ordinations of communities based on the taxonomic composition and trait structure were only weakly coupled. For example, Fig. 5 (a) shows that communities with taxonomic NMDS1 scores of around −0.2 spanned over the entire range of trait structures and, vice versa, taxonomically distinct communities were associated with similar trait structures. The taxonomic composition of communities was linked to a different set of drivers, the most important being pH, water temperature and water residence time (Fig. 5a,b). Finally, whereas from a functional point of view lakes seemed to be much less similar to each other than rivers, the opposite was true in terms of taxonomic dissimilarity (Fig. 5c,d).
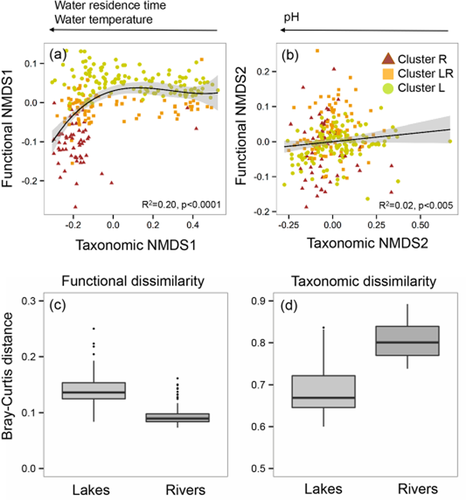
(a), (b) Comparison between the taxonomic and functional non-metric multidimensional scaling (NMDS) axes. Samples are coloured/shaded based on the three functional clusters (see Fig. 3) to show that the taxonomic composition of communities does not follow the gradient in trait structure depicted by the three functional groups. The arrows indicate the variables with the strongest correlations with both taxonomic NMDS axes (see Appendix S2). We also tried the other two possible combinations of functional and taxonomic NMDS axes (NMDS1 against NMDS2) but the correlations were even weaker and are not shown. (c), (d) Comparison between functional and taxonomic dissimilarity (Bray–Curtis distance) measured among lakes and rivers. Differences between systems were statistically significant in both cases (P < 0.0001).
Discussion
In this paper we have explored the large-scale spatial patterns in the trait structure associated with bacterial processing of DOC in boreal river and lake bacterioplankton assemblages. At the community level, broad microbial processes are the result of multiple related functions enabled by an ensemble of functional traits distributed among community members, which we term the ‘community trait structure’ (Fig. 1a). Our approach is based on exposing bacterial communities from a wide range of aquatic environments to the same set of carbon substrates under similar conditions. The exploration of the large-scale spatial variation in the resulting metabolic profiles, which we interpret as an expression of the trait structure underlying bacterial carbon processing (Fig. 1b), allows us to identify the factors that potentially shape the trait structure at the landscape level.
We are well aware of the shortcomings associated with the Biolog technique. It is important to note that we do not seek to derive actual substrate consumption rates or to identify which compounds fuel microbial activity in situ in these waters, but rather to infer from these profiles differences in the functional responses between communities that have been exposed to largely different environmental histories. While it is a fact that the incubation under a single carbon source necessarily leads to a shift in the dominance in the wells due to selective growth of certain taxa (Preston-Mafham et al., 2002), this selection operates over the existing taxa based on their respective capacities. Therefore, the pattern of substrate use does reflect a functional response of that ambient community, which is pre-determined by the distribution of the targeted functional capacities within the initial community, for example if these capacities are distributed widely among the dominant taxa or restricted to rare taxa. In this regard, although the maximum incubation time was 9 days, the metabolic profiles were recorded when the average well colour development was 0.5, which for most samples occurred within 1–4 days. This approach compensates for differences in the initial bacterial inoculum and community growth rates between samples, but this time frame also allows for functionality in rarer taxa to be expressed while maximizing the differential expression of these capacities. Moreover, as part of our own previous work with this technique, we have observed that replicate Biolog plates from the same sample yield remarkably similar and consistent metabolic profiles (see Comte & del Giorgio, 2009). All this supports the notion that these profiles are not just the result of the random growth of taxa but rather reflect intrinsic functional properties of the targeted communities.
Overall, we observed a clear segregation of the sampled sites based on regional patterns in major environmental and geographical factors across the boreal landscape (Fig. 2b), including variables such as pH, temperature or DOC concentration that have been shown to influence bacterial distribution (e.g. Lindström et al., 2005; Martiny et al., 2011; Fujii et al., 2012). This regional segregation, however, was not reflected in the large-scale spatial patterns of bacterioplankton community trait structure, which instead seemed to follow a continuum in the amount and quality of DOM largely driven by the degree of terrestrial influence (Fig. 3). In support of this observation, we identified three distinct clusters of sites based on their respective trait structure that mostly differed in the composition of their DOM pools and their degree of connectivity to land (Fig. 3b, Table 1). This greater role of environment versus geographical location in shaping functional attributes over large spatial scales was recently suggested for the molecular diversity associated with function in marine communities (Raes et al., 2011; Jiang et al., 2012), but never before for actual functions, and highlights the importance of the environment as a filter for freshwater bacterial functional capacities at the landscape scale. We acknowledge that our results provide a midsummer snap-shot and do not incorporate the potential temporal variability in carbon supply (e.g. Lambert et al., 2013), yet when the functional profiles were analysed for a subset of rivers and lakes sampled on three occasions (spring, summer and autumn, n = 132; data not shown) the same pattern was observed: irrespective of the season, rivers and lakes segregated along a cDOM gradient, highlighting the consistency and strength of the spatial patterns presented here.
Our analysis therefore suggests that, from a functional perspective, bacterial communities do not experience lakes and rivers as distinct per se, but rather as an aquatic continuum of the changing nature of the DOM that is largely driven by the degree of terrestrial influence (Fig. 3). DOM is molecularly transformed along its transit through aquatic networks (Battin et al., 2008; Fellman et al., 2014; Kothawala et al., 2014), and these changes in the quality of DOM have been shown to have a significant impact on freshwater bacterial carbon processing by shaping bacterial DOC consumption and respiration patterns (McCallister & del Giorgio, 2008; Guillemette et al., 2013; Lapierre et al., 2013; Fellman et al., 2014). The results presented here further suggest that those previously reported bacterial metabolic responses to DOM are probably mediated by major shifts in bacterial trait structure. For example, we found that the metabolic profiles associated with high-cDOM environments (cluster R) were characterized by a greater maximum average substrate respiration but a more uneven use of the substrates (Table 1), a pattern that was not associated with differences in the initial bacterial abundances or nutrient concentration (details not shown).
We also found an increase in functional divergence between communities as ecosystems become more disconnected from land, as observed by the rise in functional dissimilarity between communities inhabiting waters with less cDOM and fresher, more diverse DOM pools (Fig. 4). In this regard, the material loaded from land would seem to provide a narrower resource space for bacterial communities, and the differentiation of function along the aquatic DOM continuum may result from the diversification of the DOM pool due to local (in-lake or river) processes coupled with increases in water residence time (Kothawala et al., 2014). This differentiation of the available DOM pool as new sources and sinks develop during transit would result in a more heterogeneous resource space, in turn driving a progressive divergence in the functionalities among bacterial assemblages.
Compared with the functional patterns, lake and river communities were much more clearly differentiated in terms of taxonomic composition (anosim R = 0.59, P < 0.0001; Appendix S2), which largely changed along an aquatic continuum from the smallest headwater streams towards large rivers and lakes (details not shown). Thus, the cross-system convergence in bacterial trait structure linked to the quality of DOM was not reflected in taxonomic composition, which instead appeared to be structured by a different set of environmental and hydrological factors, such as pH (which seemed to cause a certain regional segregation; anosim R = 0.16, P < 0.0001), temperature, and water residence time (J.P.N.-G. et al. in prep.; and see Fig. 5 and Appendix S2), probably indicating a large degree of functional redundancy among taxonomically distinct aquatic bacterial communities. This lack of coherence between functional and taxonomic patterns is in accordance with previous results from studies using genomic traits (Burke et al., 2011; Barberán et al., 2012) and suggests that the exploration of functional attributes could unveil drivers of microbial performance that cannot be reconstructed from the distribution patterns of extant taxa.
Interestingly, taxonomy and function also differed in their patterns of dissimilarity, and while river assemblages were less taxonomically similar to each other than lake communities, the opposite was true for the trait structure (Fig. 5c,d). A similar pattern was recently observed in boreal systems from Sweden, where stream communities were found to be more functionally but less taxonomically similar to each other than those from lakes (Severin et al., 2014). This greater taxonomic heterogeneity among bacterial communities in rivers may be caused by the inoculation of bacteria from soils, known to be more diverse and spatially heterogeneous than in aquatic communities (Tamames et al. 2010; Torsvik et al., 2002), and which often results in a pattern of decreased diversity from headwater streams along the aquatic continuum (Crump et al., 2012; Besemer et al., 2013; C.R.-G. et al., in prep.). In our case, the more homogeneous functional profiles observed among rivers could be due to patterns in cell activity if just a small fraction of the imported bacteria are active during transport, and only when the water residence time is long enough communities do develop in response to local conditions (Crump et al., 2007, 2012). Should this be the case, terrestrial DOM would act as a strong filter of traits, by activating certain taxa that carry particular combinations of traits needed for degrading the available substrates. In contrast, the increase in resource heterogeneity along the aquatic continuum would allow the establishment of a greater diversity of active taxa, thus augmenting the core for possible combinations of traits and increasing the heterogeneity in bacterial trait structure between lakes.
In summary, we show that the distribution of bacterial functional traits associated with the processing of organic matter over large and heterogeneous landscape gradients is linked to the DOM pool to which bacteria are exposed, regardless of pronounced differences in climate, landscape and other physicochemical conditions. The observed patterns suggest that terrestrial DOM results in a relatively narrow resource space for bacterial communities, as reflected by the rather homogeneous trait structures shared by communities inhabiting high-cDOM environments. As systems become increasingly disconnected from land, there is a divergence in function among aquatic communities probably caused by the structural diversification of the available DOM pool due to local processes. The lack of coherence with the large-scale patterns in bacterial community composition suggests a high level of functional redundancy within these bacterioplankton communities, and that the spatial structuring of species and traits does not necessarily follow the same rules. As a consequence, the functional biogeography of boreal bacterioplankton communities associated with carbon processing cannot be reconstructed from the patterns in composition, which is probably also the case for other key microbial processes. This highlights the need to develop and apply proxies for other trait structures to begin building a trait-based understanding of the functioning of these microbial communities at the whole landscape scale.
Acknowledgements
We thank Annick St-Pierre, Alice Parkes and the whole CarBBAS team for their contribution to the field and laboratory components of this research, and Adam Heathcote and Beatrix Beisner for advise on analyses and critical comments. This study is part of the programme of the Carbon Biogeochemistry in Boreal Aquatic Systems (CarBBAS) Industrial Research Chair, co-funded by the Natural Science and Engineering Research Council of Canada (NSERC) and Hydro-Québec.
References
Clara Ruiz-González is a post-doctoral researcher at the Université du Québec à Montréal, Montreal, Quebec, Canada, where she focuses on the biogeography and functioning of bacterial communities from a variety of aquatic and terrestrial environments.




