Anthropogenic N deposition alters soil organic matter biochemistry and microbial communities on decaying fine roots
Abstract
Fine root litter is a primary source of soil organic matter (SOM), which is a globally important pool of C that is responsive to climate change. We previously established that ~20 years of experimental nitrogen (N) deposition has slowed fine root decay and increased the storage of soil carbon (C; +18%) across a widespread northern hardwood forest ecosystem. However, the microbial mechanisms that have directly slowed fine root decay are unknown. Here, we show that experimental N deposition has decreased the relative abundance of Agaricales fungi (−31%) and increased that of partially ligninolytic Actinobacteria (+24%) on decaying fine roots. Moreover, experimental N deposition has increased the relative abundance of lignin-derived compounds residing in SOM (+53%), and this biochemical response is significantly related to shifts in both fungal and bacterial community composition. Specifically, the accumulation of lignin-derived compounds in SOM is negatively related to the relative abundance of ligninolytic Mycena and Kuehneromyces fungi, and positively related to Microbacteriaceae. Our findings suggest that by altering the composition of microbial communities on decaying fine roots such that their capacity for lignin degradation is reduced, experimental N deposition has slowed fine root litter decay, and increased the contribution of lignin-derived compounds from fine roots to SOM. The microbial responses we observed may explain widespread findings that anthropogenic N deposition increases soil C storage in terrestrial ecosystems. More broadly, our findings directly link composition to function in soil microbial communities, and implicate compositional shifts in mediating biogeochemical processes of global significance.
1 INTRODUCTION
The microbial decay of fine root litter is a major component of the terrestrial carbon (C) cycle (Schlesinger & Bernhardt, 2013), but our understanding of the soil microorganisms mediating this biogeochemically important process is limited (Silver & Miya, 2001). Globally, the production of fine root litter accounts for ~22% of terrestrial net primary production (NPP; McCormack et al., 2015) and ~50% of plant litter entering soil (Freschet et al., 2013). Moreover, mounting evidence indicates fine root litter is the primary source of soil organic matter (SOM; Jackson et al., 2017; Rasse, Rumpel, & Dignac, 2005; Thomas, Zak, & Filley, 2012), which is the largest pool of terrestrial C (Batjes, 1996). However, it is presently unclear which ecological factors control the decay of fine roots (Hobbie, Oleksyn, Eissenstat, & Reich, 2010; Schimel & Schaeffer, 2012; Silver & Miya, 2001; Sun et al., 2018), as well as how the microbial metabolism of fine roots into SOM will be impacted by anthropogenic environmental change.
We have established that ca. 20 years of experimental nitrogen (N) deposition, which simulates a pervasive driver of global change (Galloway et al., 2004, 2008), has slowed fine root decay and increased soil C (+18%) across the geographic extent of a northern hardwood forest ecosystem in the Upper Great Lakes region (Xia, Talhelm, & Pregitzer, 2017, 2018; Zak, Holmes, Burton, Pregitzer, & Talhelm, 2008). Although experimental N deposition has not altered the production of leaf (Pregitzer, Burton, Zak, & Talhelm, 2008) or fine root litter (Burton, Pregitzer, Crawford, Zogg, & Zak, 2004), it has slowed the decay of both (Xia, Talhelm, & Pregitzer, 2017, 2018; Zak et al., 2008). Previously, we established that fine root litter accounts for 70% of lignified plant material entering soil in our experiment (Xia, Talhelm, & Pregitzer, 2015), as well as the majority of lignin-derived monomers in SOM (Thomas et al., 2012). Thus, it appears that C derived from fine roots, not leaf litter, has increased soil C storage under experimental N deposition. However, we presently do not understand how experimental N deposition has altered the community of microorganisms metabolizing fine root litter into SOM.
We previously obtained evidence that experimental N deposition has slowed lignin decay in fine root litter to a greater extent than leaf litter, a response that has occurred despite no effect of experimental N deposition on the biochemistry of fine root litter (Xia, Talhelm, & Pregitzer, 2017, 2018). This difference plausibly arises from the high lignin content of fine roots (45%) relative to leaf litter (14%; Xia et al., 2015), and because lignin content controls the long-term rate of plant litter decay (Barnes, Zak, Denton, & Spurr, 1998; Berg, 2014). Although lignified material was previously quantified as acid insoluble fraction (AIF) in our long-term experiment, which can include other recalcitrant compounds (Xia et al., 2015; Xia, Talhelm, & Pregitzer, 2017), AIF was highly predictive of lignin content in fine roots (Xia et al., 2017). Importantly, the physiological capacity to metabolize lignin varies among and within fungi and bacteria. For example, some fungal species in the class, Agaricomycetes, deploy class II peroxidase enzymes to completely oxidize lignin to CO2 (Floudas et al., 2012; Kirk & Farrell, 1987), whereas some species in the phylum, Actinobacteria, and other bacterial lineages incompletely degrade lignin into soluble phenolic compounds (Ahmad et al., 2010; Bugg, Ahmad, Hardiman, & Singh, 2011; Kirk & Farrell, 1987). In our long-term study, experimental N deposition has slowed leaf litter decay by reducing peroxidase gene expression (−73%; Zak et al., 2019), altering expressed peroxidase composition (Entwistle, Romanowicz, et al., 2018), and increasing the potential for incomplete lignin decay by bacteria (Eisenlord et al., 2013; Freedman & Zak, 2014), but it has not altered the abundance of ligninolytic fungi on this substrate (Entwistle, Zak, & Argiroff, 2018; Freedman, Upchurch, Zak, & Cline, 2016; Hassett, Zak, Blackwood, & Pregitzer, 2009). However, the concentration of lignin in fine root litter is three times greater than in leaf litter (Xia et al., 2015), and we previously found that experimental N deposition decreases the abundance of ligninolytic fungi on lignin-rich artificial substrates decaying in the field (Entwistle, Zak, et al., 2018). If experimental N deposition has also decreased the abundance of ligninolytic fungi on fine root litter, this response could explain why fine root decay has slowed to a greater extent than leaf litter. If this expectation is correct, then reduced fine root decay under experimental N deposition should be the primary source of C accumulating in soil due to experimental N deposition, which should alter SOM biochemistry by increasing the contribution of lignin-derived compounds to SOM formation.
Here, our objective was to determine if anthropogenic N deposition has altered the composition of soil microorganisms decaying fine root litter. To accomplish this, we compared the composition of fungal and bacterial communities colonizing decaying fine root litter exposed to ambient N and experimental N deposition. We also investigated the biochemical composition of SOM under ambient and experimental N deposition to determine if, by slowing the decay of fine roots, experimental N deposition has increased the concentration of lignin-derived compounds in SOM.
2 MATERIALS AND METHODS
2.1 Description of study sites
We tested the effects of experimental N deposition on the composition of microbial communities decomposing fine root litter and the biochemical composition of both fine root litter and SOM in four replicate northern hardwood forest stands in upper and lower Michigan, USA (Figure S1). Each stand contains six 30 × 30 m plots; half receive ambient N deposition (n = 3) and half have received experimental N deposition since 1994 (n = 3; ambient N + 30 kg N ha−1 year−1 as NaNO3 pellets in six equal applications during the growing season). To reduce edge effects, each plot is surrounded by a 10 m wide buffer zone that receives the same treatment as its respective plot. The forest stands are dominated by sugar maple (Acer saccharum Marsh., >80% basal area) on sandy spodosols that are Typic Haplorthods of the Kalkaska series (>85% sand). The forest floor consists of a thick Oe/Oa horizon that contains a mat of fine roots at its boundary with the A horizon. The forest stands are matched in both vegetation and soil characteristics (Burton, Ramm, Pregitzer, & Reed, 1991) and encompass the full latitudinal range of the northern hardwood ecosystem in the Upper Great Lakes region; this ~500 km distance spans gradients of ambient N deposition, mean annual temperature, and precipitation (Table S1). Thus, our experimental design allows us to generalize our findings across this important and widespread ecosystem.
2.2 Field-based decomposition experiment
To obtain fine roots for our field decomposition experiment, we collected 60 soil cores (5 cm diameter) to a depth of 10 cm in each plot, which included both Oe/Oa and A horizons (sensu Xia, Talhelm, & Pregitzer, 2018). Although these soil cores contain fine root material from both the O and A horizons, the vast majority are derived from the dense mat of fine roots that sits at the O/A horizon boundary (Xia et al., 2018; Zak, Freedman, Upchurch, Steffens, & Kögel-Knabner, 2017). We transported the cores on ice to the University of Michigan and stored them at −20°C. Sample collection was carried out in September and October 2013. We thawed the soil cores, passed them through a 2 mm sieve, retrieved first- through third-order fine roots (Pregitzer et al., 2002; Xia et al., 2015), and pooled the roots by plot. We rinsed soil from the roots and dried them at 60°C for 24 hr. We collected the three distal root orders because, as the ephemeral absorptive modules of the root network, they are morphologically similar and exhibit the highest turnover (Guo et al., 2008; McCormack et al., 2015; Xia, Guo, & Pregitzer, 2010), thus comprising the largest input of fine root C to soil.
We placed three mesh litter bags of fine roots (~2 g dry mass in each bag) at three separate positions in the same plot from which the roots originated, in their original location in the soil profile at the boundary of the Oe/Oa and A horizons (3 litter bags × 24 plots = 72 litter bags total). While it could be argued that fine roots may have decayed differently had they been incubated at the surface of the O horizon or deeper in the mineral soil, one of the few studies to test the effects of vertical location in the soil profile on fine root decay found that fine roots located in the O and A horizons of a red pine (Pinus resinosa) plantation did not decay at different rates (Li, Fahey, Pawlowska, Fisk, & Burtis, 2015). Moreover, the vast majority of fine roots in these northern hardwood forest stands are located at the boundary of the O and A horizons (Xia et al., 2018; Zak et al., 2017). Thus, we are confident that the abiotic and biotic conditions experienced by the fine roots we deployed reflected those experienced by the majority of fine root litter in these forests. We constructed each 15 × 15 cm litter bag with 300 µm polyester mesh on top and 20 µm polyester mesh on the bottom, which allowed microfauna and fungal hyphae to enter the bags, respectively (Hobbie, 2005; Xia et al., 2018). Litter bags were placed in the field in June 2014, collected after 12 months of decomposition, and immediately stored on ice. Each bag was weighed, and its contents were homogenized by hand. A subsample was removed for physical and chemical analyses, dried at 60°C for 24 hr, and the remaining material was stored at −80°C prior to microbial community analyses.
2.3 DNA isolation
To determine if experimental N deposition altered the composition of fungal and bacterial communities, we characterized these communities using ribosomal DNA (rDNA) sequence abundances. We isolated total genomic DNA from three replicate subsamples taken from each root litter bag (0.05 g fine root material per subsample) using the DNeasy Plant Mini Kit (Qiagen) following a modified manufacturer's protocol. Specifically, following chemical lysis as specified, we performed physical lysis by bead beating with four 2.38 mm stainless steel beads at 1,200 rpm for 45 s using the PowerLyzer 24 Bench Top Bead-Based Homogenizer (MoBio Laboratories). Debris was pelleted by centrifugation at 16,000 g for 5 min. After DNA extractions were completed, we verified the quality of extracted DNA with a NanoDrop 8000 Spectrophotometer (Thermo Scientific) and gel electrophoresis. We pooled replicate extractions from each litter bag and stored DNA at −80°C prior to PCR amplification.
2.4 PCR amplification, amplicon sequencing, and sequence quality control
We performed PCR amplification of fungal rDNA using the primers LROR and LR3 (Vilgalys & Hester, 1990) that target the D1–D2 region of the 28S rRNA gene, which is suitable for both taxonomic and phylogenetic analyses (Liu, Porras-Alfaro, Kuske, Eichorst, & Xie, 2012; Porter & Golding, 2012). The V1–V3 regions of the bacterial 16S rRNA gene were targeted using the primers 27f and 519r (Lane, 1991). For each gene, we performed triplicate PCR reactions for each sample using the Expand High Fidelity PCR System (Roche) and a Mastercycler ProS thermocycler (Eppendorf). PCR reaction conditions are described in Table S2. Primers contained an additional 16 bp barcode for sample multiplexing for sequencing (described below; for barcode sequences, see Table S3).
We pooled triplicate reactions and purified PCR products using the MinElute PCR Purification Kit (Qiagen). The quality of purified PCR products was assessed as described above, and we quantified DNA mass with the Quant-iT PicoGreen dsDNA Assay Kit (LifeTechnologies) and a BioTek SynergyHT Multi-Detection Microplate Reader (BioTek Instruments). Sequencing was performed at the University of Michigan DNA Sequencing Core on 16 SMRT chips with a PacBio RS II system (Pacific Biosciences) utilizing circular consensus sequencing, which achieves error rates comparable to other high-throughput sequencing platforms (Fichot & Norman, 2013; Travers, Chin, Rank, Eid, & Turner, 2010). PCR products were pooled in equal masses per sample per SMRT chip prior to sequencing. Mean amplicon lengths were 688 and 525 bp for fungal 28S and bacterial 16S, respectively. Only sequences with at least fivefold circular consensus coverage were retained.
We processed sequences using mothur v1.40.5 (Schloss et al., 2009). We removed sequences containing homopolymers >8 nucleotides in length, with average quality scores <30 using a 50-nt sliding window, an ambiguous base call, or >1 mismatch in either the barcode or primer sequence. Fungal sequences were aligned against a 28S reference alignment from the RDP LSU training set (Mueller, Balasch, & Kuske, 2014) and bacterial 16S sequences were aligned against the SILVA v132 reference alignment (Quast et al., 2013). Chimeric sequences were identified using UCHIME (Edgar, Haas, Clemente, Quince, & Knight, 2011) and removed. We clustered fungal sequences and bacterial sequences into operational taxonomic units (OTUs) at 99% and 97% sequence similarity, respectively. The most abundant sequence for each OTU was used as the representative for that OTU, and taxonomic assignments were made using the RDP classifier with the LSU training set v11 for fungi (Cole et al., 2014) and the SILVA v132 reference alignment with the naive Bayesian classifier (Wang, Garrity, Tiedje, & Cole, 2007) in mothur for bacteria. Raw sequences are available in fastq format in GenBank under the accession numbers SRR8591550 (16S) and SRR8591551 (28S).
2.5 Microbial community composition
Some fungi in the class Agaricomycetes, and some bacteria in the phylum Actinobacteria, can metabolize lignin (Floudas et al., 2012; Kirk & Farrell, 1987); thus, we tested if experimental N deposition altered the relative abundances of these two groups. We further summed sequence abundances in fungal orders and bacterial families, and compared relative abundances between the ambient and experimental N deposition treatments for orders and families that accounted for at least 1% of fungal and bacterial sequences, respectively, and exhibited a change in relative abundance of at least 20%. Furthermore, species of Agaricomycete fungi and Actinobacteria span a diverse range of autecologies (Hibbett et al., 2014; Kirk & Farrell, 1987), and it is difficult to directly interpret the functional consequences of changes in the relative abundance of these broad groups. Thus, we assessed the effect of experimental N deposition on fungal and bacterial community composition (i.e., β-diversity), using multivariate analyses at the genus and family levels, respectively. First, abundances were Hellinger-transformed to avoid subsampling biases (McMurdie & Holmes, 2013, 2014). We then performed distance-based redundancy analysis (db-RDA) on Bray–Curtis dissimilarity calculated from these abundances to visualize differences in community composition due to site and experimental N deposition. We plotted the scores for abundant (>1%) classified fungal genera and bacterial families to determine which taxa drove differences in community composition in response to experimental N deposition.
2.6 Biochemical analyses and relationships with microbial community composition
We characterized the biochemical composition of undecomposed fine roots, decayed fine roots, and SOM using pyrolysis gas chromatography-mass spectrometry (py-GC/MS). Mineral soil (0–10 cm) was obtained from each plot receiving ambient N and experimental N for biochemical analysis of SOM. We elected to characterize the biochemistry of SOM in mineral soil for four reasons. First, organic matter has rapidly accumulated (+18%) in the mineral soil of our experiment (Zak et al., 2008). Second, the lignin-derived compounds remaining in mineral soil appear to be derived primarily from fine root litter (Thomas et al., 2012), emphasizing the importance of relating microbial composition on fine root litter to the biochemistry of SOM in mineral soil. Third, we recently obtained evidence that experimental N deposition has caused an accumulation of occluded particulate organic matter in our experiment, which was hypothesized to be an accumulation of fine root-derived C (Zak et al., 2017). Finally, previous biochemical characterizations of mineral soil SOM have not detected the expected accumulation of lignin-derived compounds in response to experimental N deposition (Thomas et al., 2012; Zak et al., 2017); thus, we employed a high-resolution method (i.e., py-GC/MS) to definitively test this alternative. Dried fine root and soil samples (~1 g per sample type per plot) were ground for 6 min using a ball mill. Samples were then pyrolyzed at 600°C in quartz tubes for 20 s using a DS Pyroprobe 5150 pyrolyzer, and analyzed using a ThermoTrace GC Ultra gas chromatograph (Thermo Fisher Scientific) and ITQ 900 mass spectrometer (Thermo Fisher Scientific; sensu Pold, Grandy, Melillo, & DeAngelis, 2017). Mass spectrometry peaks were assigned to compounds using AMDIS software and a previously compiled compound library, and relative abundances for each compound were determined by dividing by the largest peak present in that sample (Grandy, Neff, & Weintraub, 2007; Grandy, Strickland, Lauber, Bradford, & Fierer, 2009; Wickings, Grandy, Reed, & Cleveland, 2011). Individual compounds were summed by their origins to determine the relative abundances of broad compound classes (i.e., aromatic, lignin, lipids, N-bearing, phenols, polysaccharides, proteins, and compounds of unknown origin). To evaluate if SOM biochemistry was related to microbial community composition on decaying fine roots, we fit vectors of compound abundances in SOM to db-RDA ordinations and overlaid vectors with a significant fit (see Statistical analyses 2.7).
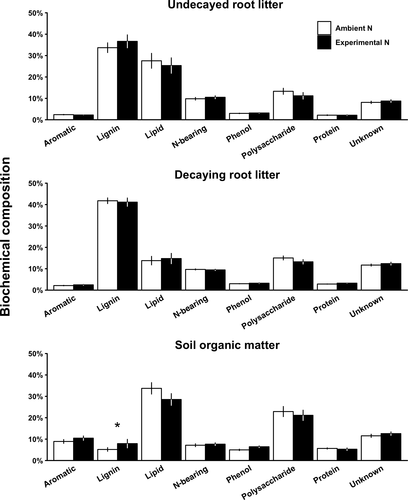
Although compounds other than lignin, such as suberin, are also important biochemical constituents of fine roots (McCormack et al., 2015), we elected to focus our study on lignin for four reasons. First, lignin dominated fine root litter biochemistry (35%–45%) in our long-term experiment based on previous findings (Xia et al., 2015, 2017) and the results we have obtained in our present study (see Figure 1). Second, the biochemical composition of lignin-derived monomers in SOM in our experiment was biochemically more similar to fine root-derived lignin than to leaf litter-derived lignin (Thomas et al., 2012), a finding that specifically implicates fine root-derived lignin as an important source of SOM. Third, the decay of AIF in fine roots (which is dominated by lignin in our long-term experiment) was reduced under experimental N deposition (Xia et al., 2017, 2018), leading us to address the mechanism by which this reduction of decay has occurred in the present study. Finally, suberin is relatively more abundant in higher order (e.g., fourth and fifth order) transport fine roots, as opposed to the ephemeral absorptive fine root modules (orders 1–3; McCormack et al., 2015) that are the focus of our present study due to their dominance of fine root turnover (Xia et al., 2010, 2015). Taken together, these lines of evidence support our focus on the microbial degradation of fine root lignin in response to experimental N deposition.
2.7 Statistical analyses
We used two-way ANOVA to test the effect of experimental N deposition, site, and their interaction on Hellinger-transformed taxon abundances (e.g., Agaricales) and log2-transformed compound abundances. Among-group means were compared using protected Fisher's least significant difference test in the agricolae package (de Mendiburu, 2017) in R. We tested the effects of experimental N deposition, site, and their interaction on community composition using two-way permutational multivariate analysis of variance (PERMANOVA; Anderson, 2001) and Bray–Curtis dissimilarity matrices calculated from Hellinger-transformed fungal genus and bacterial family abundances. PERMANOVA was implemented in the vegan package v2.5-3 (Oksanen et al., 2018) in R. PERMANOVA cannot distinguish differences in composition from heterogeneous variance; thus, we tested the homogeneity of multivariate dispersion using PERMDISP (Anderson, 2004) in vegan (“betadisper” function). A nonsignificant PERMDISP result confirms that a significant PERMANOVA test has detected a true difference in composition. Vectors for compound abundances were fit to db-RDA ordinations using the “envfit” function in vegan. Due to the broad geographic expanse of our experiment and inherent heterogeneity of the soil environment, we accepted statistical significance at α = .1. Data processing and visualization were performed using the collection of packages comprising the tidyverse v1.2.1 (Wickham, 2017) in R. Statistical analyses were performed in R v3.5.1 (R Core Team, 2018) and RStudio v1.1.453 (RStudio Team, 2018), and code for sequence processing and statistical analyses is available at https://github.com/ZakLab-Soils/N-deposition_roots.
3 RESULTS
3.1 Fine root and SOM biochemistry
Experimental N deposition did not affect the relative abundance of any compound class in undecayed or decaying fine root litter (ANOVA, p > .1; Figure 1). However, we found that experimental N deposition increased the relative abundance of lignin-derived compounds in SOM by 53% (5.2% under ambient N to 7.9% under experimental N; p = .092; Figure 1). Although this response was not highly statistically significant, it was ecologically significant due to its magnitude (>50% change), its uniformity across a large geographic expanse (site by treatment interaction, p > .1), and the rapidity with which it occurred (~20 years).
3.2 Sequence processing, OTU clustering, and taxonomic distribution
Our sequencing effort yielded 126,159 high-quality (i.e., passed filtering steps described in Materials and Methods) fungal sequences (5,257 ± 1,656 per sample; mean ± SD) and 154,135 high-quality bacterial sequences (6,422 ± 1,058 per sample). We obtained 2,071 non-singleton fungal OTUs and 5,957 non-singleton bacterial OTUs across all samples. Basidiomycota (63%) and Ascomycota (35%) represented the majority of fungal sequences. The fungal classes Agaricomycetes (57%), Sordariomycetes (11%), unclassified Ascomycota (8%), Leotiomycetes (6%), Tremellomycetes (5%), and Eurotiomycetes (5%) were most abundant. Dominant bacterial phyla included Proteobacteria (55%), Bacteroidetes (15%), Acidobacteria (10%), and Actinobacteria (7%).
3.3 Effects of experimental N deposition on microbial community composition
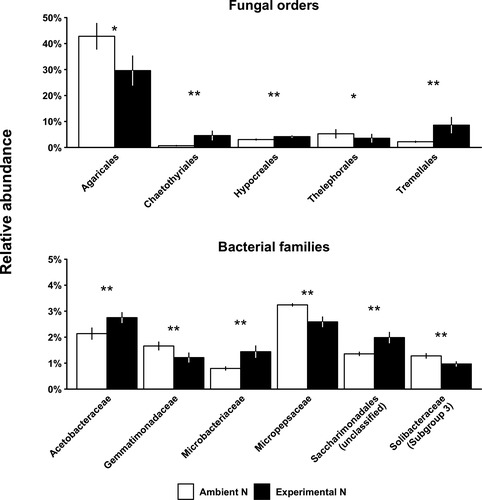
The abundance of Agaricomycetes declined (−22%, from 62.3 ± 4.8% to 48.8 ± 7.5%, mean ± SE) in response to experimental N deposition (ANOVA, p = .085; Figure S2). Similarly, experimental N deposition reduced the abundance of Agaricales (−31%; p = .059; Figure 2), the most abundant order of Agaricomycetes colonizing fine root litter. Fungal orders that responded positively to experimental N deposition did not belong to the class, Agaricomycetes. For example, experimental N deposition increased the abundance of fungal orders Chaetothyriales (+566%; p = .011), Hypocreales (+37%; p = .033), and Tremellales (+291%; p = .009; Figure 2). The responses of Hypocreales (site by treatment; p = .017) and Tremellales (p = .042) varied in magnitude, but not direction by site (Figure S3). Additionally, the relative abundance of Actinobacteria increased (+24%, from 6.5 ± 0.5% to 8.1 ± 0.7%) in response to experimental N deposition (ANOVA; treatment; p = .025), driven primarily by sites B and C (site by treatment interaction; p = .024; Figure S2). Among bacterial families, Microbacteriaceae were favored by experimental N deposition (+81%; p = .005); this response varied in magnitude by site, but not in direction (site by treatment; p = .053; Figure S3).
Experimental N deposition significantly altered the genus-level composition of fungal communities on decaying fine roots (PERMANOVA; p = .001; Figure 3a), without altering dispersion (PERMDISP; p = .17). The shift in community composition due to experimental N deposition (denoted by the “Exp. N” vector in Figure 3b) was associated with a lower abundance of the ligninolytic fungal genera Mycena and Kuehneromyces (Figure 3b). Specifically, the points labeled “Myc” and “Kue” in Figure 3b represent the loadings for these genera in the site by treatment ordination in Figure 3a; if an arrow were drawn from the origin in the ordination to a genus loading, it would represent the direction in which the abundance of that genus increased. Thus, the relative abundance of Mycena and Kuehneromyces increased in the opposite direction of the vectors representing the shift in fungal community composition due to experimental N deposition (“Exp. N”). In other words, the experimental N deposition treatment is associated with a lower abundance of these two genera. This pattern indicates that a decline in the abundance of these genera drove the significant change in fungal community composition on decaying fine roots in response to experimental N deposition.
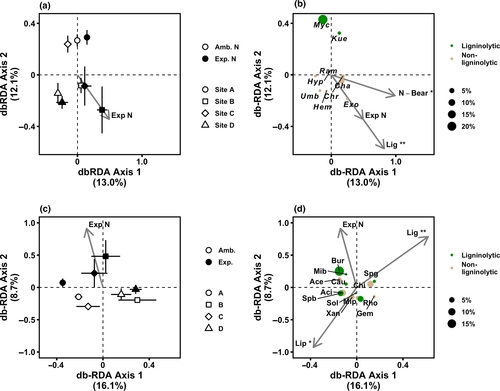
Similarly, experimental N deposition significantly altered bacterial community composition on decaying fine roots (PERMANOVA; p = .014; PERMDISP; p = .39; Figure 3c). The actinobacterial family, Microbacteriaceae, was among the bacterial families positively associated with the change in community composition due to experimental N deposition (Figure 3d). This family contains ligninolytic species (Taylor et al., 2012), which incompletely metabolize lignocellulose into soluble phenolic compounds. The effects of experimental N deposition on other bacterial families putatively involved in lignin degradation (Wilhelm, Singh, Eltis, & Mohn, 2018) were idiosyncratic. The community composition of fungal and bacterial communities differed among sites (PERMANOVA, site; p < .001). The effect of experimental N deposition was not uniform across sites for fungi or bacteria (site by treatment; p < .05). However, the significant site by treatment interaction was apparent in db-RDA ordinations (Figure 3a,c), in which clear separation occurred between communities under ambient and experimental N deposition at all sites, except site D.
3.4 Relationships between SOM biochemistry and microbial community composition
To directly link changes in SOM biochemistry with changes in bacterial and fungal community composition elicited by experimental N deposition, we fit a vector for the relative abundance of each compound class in SOM to fungal and bacterial db-RDA ordinations. We found that the shift in fungal community composition driven by experimental N deposition was significantly associated with greater relative abundances of lignin-derived compounds (r2 = .39; p = .011) and N-bearing compounds (r2 = .33; p = .022) in SOM (Figure 3b). Similarly, the change in bacterial community composition elicited by experimental N deposition was significantly related to a greater relative abundance of lignin-derived compounds (r2 = .29; p = .032; Figure 3d), although the relationship was less direct than that with fungal community composition (Figure 3b). In contrast, a lower abundance of lipids was associated with changes in bacterial community composition under experimental N deposition (r2 = .21; p = .072; Figure 3d).
4 DISCUSSION
Anthropogenic N deposition has slowed the accumulation of CO2 in the atmosphere by increasing C storage in northern forests (Keenan et al., 2017; Pan et al., 2011). Nitrogen deposition fosters this terrestrial C sink by slowing microbial litter decay and increasing SOM (Chen et al., 2018; Frey et al., 2014; Janssens et al., 2010; Pregitzer et al., 2008; Zak et al., 2008). Here, we provide evidence that anthropogenic N deposition has altered the composition of fungal and bacterial communities on decaying fine root litter by suppressing the relative abundance of ligninolytic fungi and favoring bacteria with weaker ligninolytic capacity, which plausibly explains why the decay of fine root litter has declined and soil C storage has increased in our long-term N deposition experiment (Xia et al., 2017, 2018; Zak et al., 2008). Moreover, we demonstrate that shifts in microbial community composition are significantly related to an increase in the relative abundance of lignin-derived compounds in SOM, which suggests that changes in the microbial decay of fine root litter have caused the end products of this process to accumulate as SOM to a greater extent under experimental N deposition. A recent modeling study estimated that up to 51% of C accumulating in surface soil (O and A horizons to a depth of 10 cm) in this experiment could be explained by reduced decay of fine root litter (Xia et al., 2018), and our findings shed light onto the compositional changes in microbial communities eliciting this response. Furthermore, mounting evidence suggests that anthropogenic N deposition slows fine root decay in other ecosystems (Kou et al., 2018; Sun, Dong, Wang, Lü, & Mao, 2016), and that fine root C is a primary source of SOM in general (Jackson et al., 2017; Rasse et al., 2005; Thomas et al., 2012). Thus, the microbial responses we observed here may underlie widespread findings that anthropogenic N deposition increases soil C storage in terrestrial ecosystems, including those contributing to the increasing C sink in the Northern Hemisphere that has slowed the rate at which anthropogenic CO2 has accumulated in the atmosphere (Frey et al., 2014; Janssens et al., 2010; Keenan et al., 2017; Maaroufi et al., 2015; Pan et al., 2011).
Our findings suggest that declines in the relative abundance of ligninolytic fungi have reduced fine root decay in our experiment, as well as the others detailed above. Specifically, experimental N deposition decreased the relative abundance of Agaricomycetes (−22%) and its most abundant order, Agaricales (−31%; Figure 2). Agaricomycetes contains the “white-rot” fungi, which decay lignin using class II peroxidases (Baldrian, 2008; Floudas et al., 2012; Kirk & Farrell, 1987). However, there is considerable functional diversity within the Agaricomycetes (Hibbett et al., 2014); thus, the lower relative abundance of the genera Mycena and Kuehneromyces associated with experimental N deposition (Figure 3b) is a particularly important piece of evidence we obtained. Specifically, Kuehneromyces and Mycena are genera of white-rot fungi that decay lignin using class II peroxidases (Ghosh, Frankland, Thurston, & Robinson, 2003; Hofrichter, 2002; Kellner et al., 2014; Miyamoto, 2000). Mycena were the most abundant fungi on decaying fine roots (~22% of fungal sequences overall) in our study, and were also dominant saprotrophs on decaying fine roots in other forest ecosystems (Kohout et al., 2018; Philpott, Barker, Prescott, & Grayston, 2018); thus, this genus may be important for how fine root decay responds to anthropogenic N deposition more generally. Taken together, our results clearly demonstrate that experimental N deposition is associated with a lower relative abundance of ligninolytic fungi on decaying fine roots.
In contrast, experimental N deposition favored ligninolytic bacteria and non-ligninolytic fungi. The relative abundance of Actinobacteria increased under experimental N deposition (+24%), including the family, Microbacteriaceae (+81%; Figures 2 and 3d). Experimental N deposition also increased the abundance of Saccharibacteria (+46%) and the fungal orders Chaetothyriales (+566%), Hypocreales (+37%), and Tremellales (+291%; Figure 2). These responses are likely ecologically important because ligninolytic Actinobacteria, including some Microbacteriaceae, degrade lignin to soluble phenolic compounds rather than oxidizing the polymer to CO2 (Ahmad et al., 2010; Bugg et al., 2011; Taylor et al., 2012); this is consistent with greater phenolic dissolved organic C production in our experiment (Pregitzer, Zak, Burton, Ashby, & Macdonald, 2004). Some Saccharibacteria can modify aromatic compounds, but there is no evidence to indicate they degrade lignin (Luo, Xie, Sun, Li, & Cupples, 2009). Other bacterial lineages have been implicated in lignin decay, including some that have responded to experimental N deposition (Figure 3d; Janusz et al., 2017); however, the cumulative effect of these changes in composition on bacterial lignin degradation remains to be tested. Some Hypocreales and Chaetothyriales also possess oxidases that could modify lignin (Assavanig, Amornikitticharoen, Ekpaisal, Meevootisom, & Flegel, 1992; Hölker, Dohse, & Höfer, 2002; Martinez et al., 2008; Teixeira et al., 2017), and yeasts in Tremellales dominate the late, lignin-rich stages of oak leaf litter decomposition (Voriskova & Baldrian, 2013). However, these fungal lineages lack peroxidases capable of complete lignin oxidation (Floudas et al., 2012). Together, these responses suggest that experimental N deposition has favored a microbial community with a lower capacity to degrade lignin in fine root litter.
In combination with a higher relative abundance of lignin-derived compounds in SOM, our observations specifically link changes in microbial community composition on fine root litter to the accumulation of SOM (Table S1; Pregitzer et al., 2008; Zak et al., 2008). Foremost, experimental N deposition significantly altered fungal community composition by decreasing the relative abundance of ligninolytic Mycena and Kuehneromyces, and these shifts in composition were significantly associated with a greater relative abundance of lignin-derived compounds in SOM (Figure 3a,b). Similarly, the relative abundance of lignin-derived compounds in SOM was positively related to the shift in bacterial community composition elicited by experimental N deposition (Figure 3c,d). The substantial declines in the relative abundance of ligninolytic fungi and increases in the relative abundance of bacteria with weaker ligninolytic capacity we observed (Figures 2 and 3) likely account for the reduction in fine root lignin decay (Xia et al., 2017) and mass loss (Xia et al., 2018) previously reported from our experiment, wherein fine root litter was allowed to decay in the field in an identical manner as our current study. Moreover, our findings suggest that by substantially altering the composition of microbial communities on fine roots, experimental N deposition has slowed the decay of lignin-rich fine root litter, thereby increasing the contribution of lignin-derived compounds from fine roots to SOM formation.
It is unclear why experimental N deposition decreased the abundance of ligninolytic fungi on fine root litter, whereas this response has not occurred on leaf litter in the same long-term experiment or others (e.g., Morrison et al., 2016; Morrison, Pringle, van Diepen, & Frey, 2018; Whalen, Smith, Grandy, & Frey, 2018). A reduction in the competitive ability of ligninolytic fungi on lignin-rich substrates has been proposed to explain the negative effects of experimental N deposition on ligninolytic enzyme activity and litter decay (e.g., DeForest, Zak, Pregitzer, & Burton, 2004; Entwistle, Zak, et al., 2018; Janssens et al., 2010; Morrison et al., 2018; Talbot & Treseder, 2012; Waldrop, Zak, Sinsabaugh, Gallo, & Lauber, 2004), but the mechanisms underlying putative changes in competitive ability on lignin-rich substrates are not understood. Our observation that the relative abundance of ligninolytic fungi was reduced to a greater extent on fine root litter than leaf litter could be consistent with this hypothesis, although the role of competition and its specific mechanisms are unknown. A trade-off between stress tolerance and competitive ability has recently been proposed to explain the effects of experimental N deposition on ligninolytic fungi (Morrison et al., 2018), and numerous other mechanisms involving niche differentiation and an increased efficiency of non-ligninolytic fungi have also been suggested (e.g., Talbot & Treseder, 2012). Our findings, including the relationships between microbial composition and other components of SOM (e.g., N-bearing compounds and lipids; Figure 3), emphasize the need to understand whether biotic interactions influence how experimental N deposition alters microbial community composition. For example, the distinction between these putative competition-mediated changes in composition and physiological responses (i.e., downregulated peroxidase transcription) would be represented differently in mechanistic ecosystem models (Allison, 2012; Hawkes & Keitt, 2015; Treseder et al., 2012). At present, these competitive processes are speculative and their mechanisms are not understood; a mechanistic understanding of these interactions will facilitate their extension to the effects of anthropogenic N deposition on fine root decay and soil C storage in other ecosystems.
The fact that experimental N deposition did not alter the biochemical composition of fine roots after one year of decay (Figure 1), and that it did increase the lignin content of SOM (Figures 1 and 3), indicates that the changes in microbial community composition we documented have functional implications during the later stages of fine root decay (i.e., beyond 1 year). Several pieces of evidence from our long-term experiment are consistent with this expectation. For example, based on the decay of identical fine root litter in identical litter bags, there was no effect of experimental N deposition on the mass loss (Xia et al., 2018) or biochemistry (Xia et al., 2017) of fine root litter after one year of decay. However, experimental N deposition significantly increased the mass of fine root litter remaining after 3 years of decay (Xia et al., 2018) due to a reduction in the decay of lignin (Xia et al., 2017). These reductions in the later stages of fine root decay align with the accumulation of lignin-derived compounds in SOM revealed in our current study (Figures 1 and 3). An important assumption is that the changes in microbial community composition we observed after one year persist to later stages of decay, thereby decreasing the loss of lignin and overall mass loss of fine root litter. Although this assumption remains to be tested, our findings clearly suggest that changes in microbial community composition (Figures 2 and 3) have slowed the decay of lignin in fine root litter (Xia et al., 2017, 2018), thereby increasing the amount of lignin-derived compounds from fine root litter in SOM (Figures 1 and 3).
The biochemical changes in SOM we observed may explain how experimental N deposition has increased the physical protection of SOM by mineral occlusion, as we have previously reported (Zak et al., 2017). Although relatively unmodified lignin is not thought to remain in long-term pools of SOM (Grandy et al., 2007), it can be stabilized through the adsorption of dissolved organic matter to mineral surfaces, or the physical occlusion of particulate litter by clay and silt particles in microaggregates (Cotrufo et al., 2015; Lehmann & Kleber, 2015). In our experiment, experimental N deposition has not altered the amount of C in the highest density soil fraction (>1.8 g/cm) that represents mineral-adsorbed SOM; however, it has increased mineral-occluded particulate SOM, which indicates greater physical protection of litter fragments in microaggregates (Zak et al., 2017). Previous analyses have revealed no effect of experimental N deposition on SOM biochemistry or other factors involved in aggregate formation (Thomas et al., 2012; Zak et al., 2017). However, it is plausible that a reduction in the microbial decay of fine root litter has increased the amount of time a given mass of fine root fragments remain in contact with soil particles, thereby fostering their occlusion (Cotrufo et al., 2015). Although this mechanism remains to be directly tested, our results suggest that reduced microbial decay of fine root litter may increase the physical stabilization of fine root material in microaggregates, which could influence the longevity of the terrestrial C sink.
A reduction in soil pH has recently been proposed as the primary mechanism by which experimental N deposition decreases the microbial decay of plant litter and increases soil C storage (Averill & Waring, 2018, and references therein); however, our findings provide a distinct and novel mechanism that is independent of soil pH. For example, experimental N deposition induced Mn-limitation in soils receiving experimental N deposition in an oak-dominated forest in New England, likely due to enhanced leaching of Mn from soils at low pH (Whalen et al., 2018). Since the late stages of litter decay (dominated by lignin degradation) occur more rapidly when Mn concentrations are high (Berg, 2014), likely due to the role of Mn as a diffusible redox mediator for ligninolytic manganese peroxidase enzymes (Hofrichter, 2002), pH-induced Mn-limitation was thought to explain reduced rates of litter decay (Whalen et al., 2018). Additionally, experimental N deposition could reduce microbial activity due to the direct negative effects of low pH on microbial physiology (Averill & Waring, 2018). However, soil pH does not differ among sites in our long-term experiment (Table S1) nor has experimental N deposition decreased soil pH (4.5 ± 0.25 under ambient N conditions and 4.7 ± 0.32 under experimental N conditions; Eisenlord & Zak, 2010). Thus, neither Mn-limitation nor the direct negative effects of low soil pH on microbial activity explain reductions in fine root decay in our experiment. Instead, our findings suggest a pH-independent mechanism, in which the decreased abundance of highly ligninolytic fungi and increased role for less complete bacterial lignin degradation have slowed the decay of fine root litter.
In summary, we demonstrated that over 20 years of experimental N deposition has reduced the relative abundance of ligninolytic fungi and increased that of ligninolytic bacteria on decaying fine roots, which plausibly explains how fine root decay has slowed and SOM has accumulated in our study (Xia et al., 2017, 2018; Zak et al., 2008). Furthermore, we found that an accumulation of lignin-derived compounds in SOM was significantly related to changes in microbial community composition on decaying fine root litter, particularly a decline in the relative abundance of ligninolytic fungi. Together, this evidence suggests that by altering microbial community composition on fine root litter, which is the dominant source of lignified plant material to soil, experimental N deposition has caused an accumulation of root-derived C as SOM. It is important to point out that fine root litter may account for a smaller proportion of lignin-derived compounds that enter soil in forest ecosystems dominated by species with higher leaf litter lignin concentrations (e.g., Quercus, Pinus) than sugar maple. Nonetheless, our findings unite a growing body of evidence that experimental N deposition enriches SOM in compounds that are abundant in fine roots (e.g., lignin and suberin; Frey et al., 2014; Grandy, Sinsabaugh, Neff, Stursova, & Zak, 2008; van den Enden et al., 2018; Wang et al., 2019) with the changes in microbial composition that are responsible for their accumulation. To better understand how experimental N will modify terrestrial C storage and mediate climate under future rates of anthropogenic N deposition (Galloway et al., 2004, 2008), we must explicitly test ecological mechanisms (e.g., putative competitive interactions) that may alter microbial community composition and slow fine root decay, as well as better understand how the altered products of fine root decomposition are stabilized into SOM. Taken together, our findings link the composition and function of microbial communities, as well as highlight the role of compositional shifts in mediating biogeochemical processes of global significance.
ACKNOWLEDGEMENTS
We sincerely thank Kimberly Martin, Remy Long, Kathryn Kelley, Allison Sharrar, Kerri Metz, and Sisimac Duchicela for their assistance with fine root processing and litter bag construction. We thank Zachary Freedman and Karl Romanowicz for their assistance with soil collection. We also thank Peter Pellitier, Deborah Goldberg, Inés Ibáñez, Timothy James, the James lab, and the plant ecology discussion group at the University of Michigan for their insightful comments on previous versions of this manuscript. This work was supported by funding from the United States Department of Energy Biological and Environmental Research program. The maintenance of our long-term N deposition project is supported by the National Science Foundation Long-Term Research in Environmental Biology Program.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHOR CONTRIBUTIONS
DRZ designed the study. RAU, SOS, and WAA optimized and performed laboratory analyses. ASG performed biochemical analyses. WAA analyzed the data and wrote the first draft of the manuscript. DRZ, RAU, SOS, and ASG provided significant feedback on later versions.




