Undiagnosed rare disease clinic identifies a novel UBE3A variant in two sisters with Angelman syndrome: The end of a diagnostic odyssey
Rebecca Bruns, Khurram Liaqat, Erin Conboy, and Francesco Vetrini contributed equally to this study.
Abstract
Angelman syndrome (AS, MIM #105830) is a neurodevelopmental disorder characterized by severe intellectual disability, profound developmental delay, movement or balance problems, an excessively cheerful disposition, and seizures. AS results from inadequate expression of the maternal UBE3A gene (MIM #601623), which encodes an E3 ligase in the ubiquitin-proteasome pathway. Here we present the case of two sisters with features consistent with AS who had negative methylation analyses. An autism/intellectual disability expanded panel revealed a maternally inherited novel UBE3A (NM_001354506.2) variant c.2443C>T p.(Pro815Ser) in both patients that was initially classified as a variant of uncertain significance. The patients were enrolled in Indiana University's Undiagnosed Rare Disease Clinic (URDC) to further investigate the variant. Additional data, including deep phenotyping, familial segregation analysis, and in silico studies, suggest that the variant is likely pathogenic. 3D modeling studies based on the available crystal structure revealed that the Pro815Ser variant can introduce more flexibility into the protein and alter its enzymatic activity. Recent literature confirms the pathogenic nature of the variant. Reanalysis of the UBE3A variant has heightened existing knowledge of AS and has offered this family an end to their diagnostic odyssey.
1 INTRODUCTION
Angelman syndrome (AS) is characterized by developmental delay, severe speech impairment, behavioral features including frequent smiling or laughter, and movement problems such as ataxia. Seizures, abnormal and characteristic EEGs, dysmorphic facies, feeding difficulties, and altered sleep patterns may be seen.1 No disease-modifying therapies are currently approved; however, antisense oligonucleotide therapy may change future treatment strategies.2 AS results from the loss of function of the UBE3A gene that lies within the AS critical region 15q11.2–q13, spans approximately 120 kb of genomic DNA, and contains 16 exons.3
The UBE3A gene (UBE3A; MIM #601623) encodes a HECT (homologous to the E6-AP carboxyl terminus)—type of E3 ubiquitin protein ligase that ubiquitinates substrate proteins, generally leading to their degradation by the 26S proteasome. Within the HECT domain, an N-lobe attaches to ubiquitin, while the C-lobe creates an isopeptide bond that catalyzes ubiquitin transfer.4 The C-terminal HECT domain harbors the catalytic cysteine residue that is essential for transferring the ubiquitin to either itself (auto-ubiquitination) or to substrate proteins.5, 6 The ubiquitinated substrate is marked for degradation, and this regulation influences several signaling pathways. UBE3A plays a role in excitatory synapse development and functions as a transcriptional co-activator.7, 8 In mature neurons of the central nervous system, the paternal allele is imprinted.9, 10
De novo maternal UBE3A deletions, missense changes, uniparental disomy, and imprinting center defects can each lead to the loss of maternal UBE3A expression and result in AS. Methylation studies are first line testing for AS. If a methylation test is negative, and AS continues to be a differential diagnosis, gene sequencing can be used to identify UBE3A variants.11
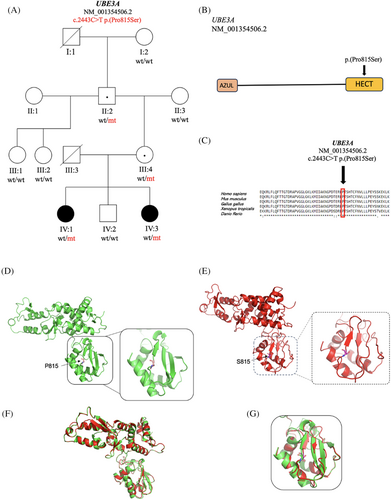
We report on two sisters who presented with features consistent with AS. Each sister had a negative methylation analysis but was subsequently found to have a maternally inherited UBE3A variant, c.2443C>T p.(Pro815Ser) in the HECT domain (Figure 1B), that was initially classified as a variant of uncertain significance (VUS). To further investigate this variant, the patients were enrolled in Indiana University's Undiagnosed Rare Disease Clinic (URDC), a multidisciplinary program with the aim to provide families affected by rare genetic diseases with diagnoses.12
2 RESULTS
2.1 Clinical presentations
In this report, we describe two sisters who presented to the URDC at ages nine (Sister 1 – IV:1) and 13 (Sister 2 – IV:3) years due to high suspicion of a genetic disorder and inconclusive genetic test results (Figure 1A). Each sister had global developmental delay, absent speech, intellectual disability, and epilepsy. They displayed autistic behaviors, stereotypy, gait ataxia, truncal hypotonia, limb hypertonia, feeding difficulties, and widely spaced teeth. Sister 1 had a history of Chiari malformation requiring three decompression surgeries, hydrocephalus requiring a VP shunt, a tethered cord requiring release, respiratory failure requiring tracheostomy, nonconvulsive status epilepticus, and obstructive sleep apnea. Sister 2 had history of respiratory failure, epileptic encephalopathy, exotropia, recurrent hand flapping, excessive hunger, autism spectrum disorder, and cerebral palsy.
2.2 Genetic testing and variant classification
The patient's family enrolled the proband (Sister 1 – IV:1) in the URDC at Indiana University School of Medicine (IUSM) because a prior molecular diagnosis had not been reached. In both patients, standard chromosome analysis, chromosomal microarray analysis (CMA), and AS methylation analyses were all negative. Additionally, myotonic dystrophy repeat analysis and PHOX2B sequence analysis were performed in Sister 1 and showed normal results. In 2018, an autism/intellectual disability expanded panel test with 2562 genes was ordered for Sister 1 as a proband and Sister 2 as a family member. In both sisters, this testing identified a variant (c.1616-4 and 1616-3delTT) in the DDX3X gene and a missense variant c.2443C>T p.(Pro815Ser) in the UBE3A gene (Table 1). Both variants were initially classified as VUS by the clinical lab (GeneDx). The DDX3X variant was determined to be of less clinical significance than the UBE3A variant because it does not fully explain the patients' phenotypes, and the same variant has been observed in multiple individuals in the gnomAD database.
| Gene | Chromosome | HGVS DNA reference | HGVS protein reference | Predicted effect | In silico predictions | Genotype | Pathogenicity | Parent of origin |
|---|---|---|---|---|---|---|---|---|
UBE3A NM_001354506.2 |
15q11.2 | c.2443C>T | p.(Pro815Ser) | Missense substitution | MUT Assessor: Hi (4.36); SIFT: Damaging (0); Revel: Deleterious (0.92); DANN: Deleterious (1); MetaLR: Deleterious (0.98) | Heterozygous | Likely pathogenic | Maternal |
The URDC obtained additional information from the patients and their family members, including detailed phenotypes and medical histories. Both the patients' asymptomatic mother and maternal grandfather tested positive for this variant (Figure 1A). The UBE3A variant c.2443C>T p.(Pro815Ser) was reclassified from a VUS to likely pathogenic following a reanalysis of the genetic data based on all available evidence. This UBE3A variant is absent in gnomAD and predicted to be damaging by various in silico tools including Mutation Assessor, SIFT, Mutation Taster, FATHMM, DANN, MetaLR, and Revel (Table 1). The Pro815 residue is highly conserved across species (Figure 1C). Variant classification was performed according to guidelines from the American College of Medical Genetics and Genomics13 and Clingen UBE3A/Angelman syndrome working group guidelines.14 The UBE3A variant was not found in the GnomAD control population (PM2_Supporting). Segregation analysis is consistent with the expected inheritance for a paternally imprinted gene as in AS (PP1_moderate). The phenotypes of our patients are consistent with AS (PP4_moderate). While Pro815 is near the known catalytic domain Cys820, evidence confirming that Pro815 itself resides within a functional domain has not been established; therefore, it is unclear if the criteria have been met for PM1 (Table 2). Finally, substitutions in the same location, Pro815His and Pro815Arg, have been functionally characterized and determined to lead to loss of function15 (PM5_moderate). Additionally, since all the in silico tools predict a damaging effect on the protein, PP3_supporting applies.
| PP4_Supporting | PP1_Moderate | PM2_Supporting | PP3_Supporting | PM5_(Moderate or strong)a | PM1 | PP2 |
|---|---|---|---|---|---|---|
| Phenotype | Segregation pattern | Absent in control population | Revel score >0.7 | Same amino acid substitution reported (two different substitutions). | Located in functional domain Unclear – very close to Cys820, which is in functional domain |
Missense variant in a gene with a low rate of benign missense variants—does not apply here |
- a Criteria can be weighted to different degrees based on clinical judgment and opinion from expert panels.
2.3 Three-dimensional modeling
The x-ray structure of ubiquitin-protein ligase E3A (PDB ID: 1C4Z) was retrieved from the Protein Data Bank. Energy minimization was performed. Then, the structure for the Pro815Ser variant was developed by replacing the proline residue with the serine residue. The structure was refined using the same energy minimization methodology as the wild-type structure. The refined structures were used to study the effect of the variant on the protein structure. PYMOL (The PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC) was used for structural visualization and image processing. In the native protein structure, the proline residue, which contains a cyclic side chain, confers rigidity to the loop region (Figure 1D). The Pro815Ser variant (Figure 1E) can introduce more flexibility into the protein structure and alter its folding, as seen in the superposed structures of wild-type and mutant proteins (Figure 1F). Loss of the beta-sheet can be attributed to the variant and can be seen in Figure 1G.
3 DISCUSSION
In this report, we describe two sisters diagnosed with AS carrying a novel UBE3A missense variant c.2443C>T p.(Pro815Ser). AS is caused by a deletion of the entire UBE3A gene in about 70%–75% of the cases, but ∼5% of the patients possess non-truncating missense variants that result in small changes in the amino acid sequence of the UBE3A protein.16, 17 The majority of AS-linked missense variants cause loss of nuclear UBE3A.18 UBE3A is widely expressed in various tissues, including the human fetal brain and adult frontal cortex. The UBE3A gene resides within the human 15q11.2-q13.3 locus that is parentally imprinted in neurons, leading to the non-Mendelian inheritance patterns of human neurodevelopmental disorders.19 AS is an autosomal dominant genetic disease secondary to paternal imprinting. Therefore, AS is primarily characterized by dysfunction of the maternal UBE3A allele.20
The novel heterozygous variant c.2443C>T p.(Pro815Ser) of the UBE3A gene found in this family was considered the cause of the phenotype of these two sisters with AS. The mother of both patients carried this variant but did not exhibit the symptoms of AS. Following verification of the mother's pedigree, this variant was found to be inherited from the maternal grandfather of the patients. The maternal great grandmother did not carry UBE3A variant. The maternal great grandfather (I:1) of both affected sisters (IV:1, IV:3) had died, so we cannot conclude whether the affected sisters' grandfather inherited the UBE3A variant from his father or if it originated in him (Figure 1A). Since the locus of the UBE3A mutant variant in both sisters was inherited from the mother, it was inferred that both sisters inherited the pathogenic UBE3A variant following a paternal imprinting. AS is a rare condition with several underlying genetic mechanisms; thus, it is imperative that researchers and clinicians collaborate to disseminate and analyze data.21 With a more thorough understanding of the pathophysiology and molecular underpinnings of AS, researchers may be able to offer promising interventions that can improve outcomes for individuals with UBE3A variants. Furthermore, we observed the Chiari malformation in the affected individual (IV:1) in this family that is not a common feature in the patients of AS.
The c.2443C>T p.(Pro815Ser) missense variant in the UBE3A gene is located in the C-lobe of UBE3A's HECT domain adjacent to the beta strand S9. Proline has a cyclic structure important for establishing the angle required in beta turns. The amino acid change may impair the ability of the UBE3A protein to fold, and improper 3D structure could hinder the catalytic function of the nearby cysteine residue Cys820.5 Different amino acid changes in nearby residues of Pro815 have been shown to have deleterious effects on nuclear localization, catalytic function, or protein folding.17 Substitutions in the same location, Pro815His and Pro815Arg, have been reported and were determined to lead to loss of function.15 Furthermore, it is evident from the three-dimensional modeling of UBE3A variant p.(Pro815Ser) that the change of amino acid from Proline to Serine at position 815 causes a conformational change in the protein (Figure 1D–G). The amino acid residue Pro815 has a significant role in structural confirmation UBE3A protein. Hence, we hypothesized that substitution of amino acid from Proline to Serine at position 815 in UBE3A caused AS in both patients (IV:1 and IV:3).
In summary, we report a novel pathogenic missense variant UBE3A in two affected sisters who presented to the URDC with epilepsy, global developmental delay, and dysmorphic facial features. The variant c.2443C>T p.(Pro815Ser) in UBE3A has been reclassified from a VUS to likely pathogenic. Following enrollment in the URDC, a multifaceted approach was applied that included variant reanalysis by clinical genomic scientists using an AI-driven platform, population and segregation studies, in silico predictions, and 3D modeling. Periodic variant reassessment, including a literature search, yielded experimental evidence supporting the pathogenicity of the UBE3A variant, ultimately ending this family's diagnostic odyssey.
ACKNOWLEDGMENTS
We extend our deep appreciation to the two sisters and their family for participating in this study. Thank you to Lili Mantcheva for assisting with IRB requirements.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
The research project used medical health information and specimens that had been collected as part of an ongoing research study at the Undiagnosed Rare Disease Clinic at Indiana University. Written informed consent was obtained from participants and/or their legal guardians for the collection, research use, and storage of the specimens according to the protocol approved on July 1, 2020, by the Indiana University Institutional Review Board (IRB# 2005902680).
Open Research
DATA AVAILABILITY STATEMENT
The UBE3A variant have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) by GeneDX under the accession number VCV000546446.2.




