Multi-omic analysis of Tyrophagus putrescentiae reveals insights into the allergen complexity of storage mites
Angel Tsz-Yau Wan, Qing Xiong and Xiaojun Xiao contributed equally to this work.
Summary Box
- A multi-omic analysis reveals the highly complex allergen profile of Tyrophagus putrescentiae.
- This comprehensive analysis of Tyrophagus putrescentiae can improve the component-resolved diagnosis of mite allergy.
Tyrophagus putrescentiae, commonly referred to as the mould mite or cheese mite, is especially well-known as a storage mite that causes human allergic diseases.1 However, it has fewer reported allergen groups compared to the house dust mites of the Dermatophagoides genus in the WHO/IUIS allergen nomenclature database.2, 3
In this omics era, multiple genome-based approaches have been boosting our understanding of the medically important mites.4-6 Using the mite allergens reported in the WHO/IUIS database as reference genes, 37 allergen groups (up to group 42) were predicted in the genome of T. putrescentiae6 and composed an allergen profile encompassing up to 85 predicted genes (Table S1). Unlike the allergen gene expression of D. farinae and D. pteronyssinus,5 the group 1 allergens (cysteine proteases) of T. putrescentiae were expressed at low levels, while the homologue of group 13 allergen, pTyr p 13.0201, exhibited the highest expression level (Figure 1A). To evaluate the IgE-binding reactivity of novel allergens, recombinant proteins were cloned, expressed and assessed by ELISA with T. putrescentiae-sensitized patient sera (Table S2). Five proteins, rTyr p 6.0101, 9.0101, 18.0101, 20.0101 and 26.0101, were suggested to be novel allergens by ELISA experiments with 11.1%, 22.2%, 11.1%, 44.4% and 50.0% positive rates, but with low IgE levels. Additional information about study methods and findings is available in the following repository https://zenodo.org/record/8429480.
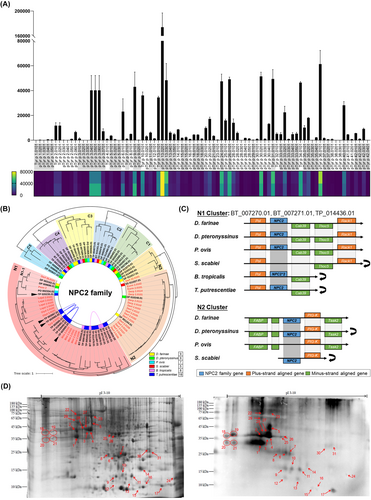
Group 2 allergens belong to the Niemann-Pick protein type C2 (NPC2) family and are considered the major allergens of storage mites including T. putrescentiae.7, 8 We identified up to six homologues of Tyr p 2, including the two homologous genes (Tyr p 2.0101 and 2.0201) which shared identical sequences, and as high as 99.3% identity with the reported Tyr p 2.
To investigate the NPC2 family, encompassing group 2, 22 and 25 allergens of mites, we gathered the genes of six astigmatic mites. The resultant phylogenetic tree revealed eight distinct clusters, named N1-3 and C1-5 (Figure 1B). All reported allergen genes were in Cluster N1-3, while none of the genes in Cluster C1-5 were reported to be allergens. Cluster N1 contained Blo t 2, Tyr p 2, Lep d 2 and Gly d 2 and Der f 35, as well as Pso o 2 (UniProt ID: Q965E2) of Psoroptes ovis. Cluster N2 covered group 2 allergens of house dust mites including Der f 2, Der p 2 and Eur m 2, while Cluster N3 contained Der f 22. The gene synteny alignment (Figure 1C) suggested that the NPC2 gene of S. scabiei in N1 decayed, while that of B. tropicalis was tandemly duplicated. Cluster N2 is proposed to be unique in psoroptid mites, but decayed in P. ovis, while a 38-aa insertion was identified in the N-terminus of SS_011027.01 in S. scabiei.
Proteomic identification was performed using the pooled sera of allergy patients (Figure 1D). In total, 31 protein spots of T. putrescentiae bound by IgE underwent peptide sequencing by MALDI-TOF mass spectrometry (Table 1). A range of our identified allergens could be found in the spots, including members of Tyr p 1, 2, 3, 8, 10, 13, 20, 21, 25, 28 and 39. Among them, the best-matched homologues in the spots included Tyr p 2.0101, Tyr p 10.0101, pTyr p 20.0101, pTyr p 21.0101, pTyr p 25.0101, pTyr p 28.0101 and Tyr p 34.0101. PTyr p 21.0101 was matched by four spots, while Tyr p 10.0101, pTyr p 20.0101 and pTyr p 25.0101 were each found in two spots. Some genes were suggested as allergen homologues but shared relatively low identity with the reported allergens, so they were labelled ungroup allergens (Table 1). However, in Tyr p 2, Tyr p 2.0101/Tyr p 2.0201, pTyr p 2.0601 and an ungrouped Tyr p 2 (gene locus: TP_020235.01) were matched. For Tyr p 28, pTyr p 28.0101, pTyr p 28.0201 and an ungrouped Tyr p 28 (gene locus: TP_006940.01) were identified.
| ID on 2D gel | Accession | Allergen | Protein | Score | MW (kDa) | pI | No. of peptides | Coverage (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | TP_005966.01 | Tyr p 10.0101 | Tropomyosin | 334.36 | 33 | 4.78 | 6 | 21.8 |
| 2 | TP_005966.01 | Tyr p 10.0101 | Tropomyosin | 265.73 | 33 | 4.78 | 4 | 14.8 |
| 6 | TP_010799.01 | pTyr p 25.0101 | Triosephosphate isomerase | 674.93 | 26.8 | 6.13 | 6 | 41.3 |
| 8 | TP_010799.01 | pTyr p 25.0101 | Triosephosphate isomerase | 610.06 | 26.8 | 6.13 | 6 | 41.3 |
| TP_007838.01 | pTyr p 3.0701 | Trypsin | 57.33 | 29.9 | 7.1 | 2 | 7.7 | |
| 9 | TP_004226.01 | – | Sod1 Superoxide dismutase [Cu-Zn] | 576.51 | 15.6 | 5.92 | 5 | 51 |
| 10 | TP_003565.04 | pTyr p 2.0601 | NPC2 family | 51.72 | 15.1 | 6.99 | 1 | 7.7 |
| 11 | TP_003565.04 | pTyr p 2.0601 | NPC2 family | 354.79 | 15.1 | 6.99 | 4 | 41.5 |
| TP_004226.01 | – | Sod1 Superoxide dismutase [Cu-Zn] | 115.71 | 15.6 | 5.92 | 3 | 23.2 | |
| TP_014856.01 | – | Unknown | 42.84 | 24.9 | 6.93 | 1 | 6.2 | |
| 13 | TP_020235.01 | Ungrouped Tyr p 2a (44.0%, CAA73221) | NPC2 family | 60.89 | 15.6 | 7.1 | 2 | 17.1 |
| 15 | TP_004518.01 | – | FK506-binding protein | 296.13 | 11.5 | 6.72 | 4 | 40.7 |
| TP_014185.01 | pTyr p 21.0101 | Unknown | 180.4 | 15.3 | 7.83 | 3 | 30.4 | |
| 16 | TP_014185.01 | pTyr p 21.0101 | Unknown | 538.38 | 15.3 | 7.83 | 6 | 46.4 |
| 17 | TP_011560.01 | Ungrouped Tyr p 13a (37.4%, AAU11502) | Fatty acid-binding protein | 392.65 | 17.5 | 8.61 | 5 | 42.9 |
| TP_003565.02/TP_003567.01 | Tyr p 2.0101/Tyr p 2.0201 | NPC2 family | 184.57 | 14.8 | 8.51 | 2 | 27.7 | |
| 18 | TP_014185.01 | pTyr p 21.0101 | Unknown | 161.6 | 15.3 | 7.83 | 3 | 30.2 |
| 19 | TP_014185.01 | pTyr p 21.0101 | Unknown | 46.76 | 15.3 | 7.83 | 1 | 10.1 |
| 20 | TP_004953.01 | – | Unknown | 132.75 | 34.2 | 9.96 | 3 | 17.9 |
| TP_005006.01 | – | Unknown | 61.66 | 34.2 | 6.9 | 2 | 11.5 | |
| 23 | TP_011350.01 | pTyr p 28.0201 | Heat shock protein 70 | 267.96 | 71.5 | 5.29 | 4 | 8.1 |
| TP_011342.01 | pTyr p 28.0101 | Heat shock protein 70 | 141.09 | 122 | 5.33 | 3 | 3.2 | |
| TP_006940.01 | Ungrouped Tyr p 28a (51.3%, AOD75395) | Heat shock protein 70 | 65.45 | 74.4 | 5.64 | 2 | 3.8 | |
| 24 | TP_001283.01 | – | Nucleoside diphosphate kinase A1 | 280.29 | 23.3 | 8.32 | 5 | 24.2 |
| TP_006307.01 | – | HEXBP DNA-binding protein HEXBP | 173.74 | 14.1 | 8.26 | 3 | 40.8 | |
| TP_020843.01 | – | CYPA Peptidyl-prolyl cis-trans isomerase | 81.44 | 28.8 | 10.08 | 1 | 5.3 | |
| TP_005620.02 | Tyr p 34.0101 | Troponin C | 52.78 | 17.7 | 4.04 | 1 | 9.8 | |
| 25 | TP_001138.03 | pTyr p 20.0101 | Arginine kinase | 421.42 | 40.1 | 6.23 | 5 | 21.7 |
| TP_009769.01 | – | FBPA Fructose-bisphosphate aldolase | 84.84 | 38.7 | 6.55 | 3 | 14.3 | |
| TP_008599.01 | Ungrouped Tyr p 1a (28.2%, ABM53753) | Cysteine protease | 69.95 | 39.5 | 5.9 | 2 | 10 | |
| 26 | TP_001138.03 | pTyr p 20.0101 | Arginine kinase | 427.58 | 40.1 | 6.23 | 5 | 21.7 |
| TP_009769.01 | – | FBPA Fructose-bisphosphate aldolase | 71.34 | 38.7 | 6.55 | 2 | 6.6 | |
| 29 | TP_007523.01 | – | pdi-2 Protein disulfide-isomerase 2 | 236.42 | 54.9 | 4.95 | 4 | 10.5 |
| TP_006736.01 | – | ATPsynbeta ATP synthase subunit beta, mitochondrial | 99.41 | 56.1 | 5.13 | 3 | 8.4 | |
| 31 | TP_013474.01 | – | Gpx5 Epididymal secretory glutathione peroxidase | 139.51 | 25.8 | 7.12 | 2 | 9.7 |
| TP_014174.01 | Ungrouped Tyr p 8a (50.7%, AGG10560) | Glutathione S-transferase | 103.13 | 24.9 | 8.95 | 2 | 12.7 | |
| TP_019125.01 | – | Cuticle protein 16.8 | 87.72 | 25.6 | 8.92 | 1 | 11.6 |
- a These genes were suggested as allergen homologues but shared relatively low identity with the reported allergens so that they were not listed in Figure 1A. The identity percentage and the GenBank accession of the reference allergen were noted in the bracket.
The ungrouped Tyr p 1 (gene locus: TP_008599.01) was identified and shared only 28.2% identity with the reported Tyr p 1 (Table 1). This cysteine protease homologue (gene locus: TP_008599.01) was estimated to be expressed over two hundred times more than the best-matched Tyr p 1.0101 and lower than 50% of that of pTyr p 3.0401 (Figure 1A). Similarly in Tyr p 13, the ungrouped homologue (gene locus: TP_011560.01) was expressed at over 50% higher levels than the highly expressed pTyr p 13.0201. Therefore, we proposed that the gene expression level was a crucial factor in proteomic identification. In addition, an ungrouped homologue of Tyr p 8 (gene locus: TP_014174.01) was identified, but not the in-silico-predicted pTyr p 8.0101 (GenBank accession: AGG10560). Other proteins such as the FK506-binding protein (gene locus: TP_004518.01) were identified in one spot (Table 1). The FK506-binding protein was assessed to be 22.2% and positive in four patient sera. The pooled sera were found to test positive for D. pteronyssinus, while their status for T. putrescentiae remained unknown. This raises concerns about potential cross-reactivity. However, it is important to note that a significant limitation of this study is the absence of immunological investigations of these allergens.
In comparison to previous omics studies focused on T. putrescentiae allergens,9, 10 our research integrates multi-omic data and highlights the presence of multiple allergen homologues in T. putrescentiae genome. Through our multi-omic approaches, a comprehensive allergen profile of T. putrescentiae was revealed and the integrative analysis provides a systematic understanding of the allergen complexity of storage mites.
AUTHOR CONTRIBUTIONS
ATYW, QX, and XX designed the experiments, analyzed the data, and wrote the manuscript. KFKA, SWJ, BSHW, MW, QC, CSHF, SMN performed the experiments and analyzed the data. FTC, BS, TFL and KYJ provided the resources. XL and SKWT supervised the study and revised the manuscript.
ACKNOWLEDGEMENTS
We would like to thank the staff members in the Shenzhen Key Laboratory of Allergy for the mite culturing work.
FUNDING INFORMATION
This work was made possible by grants: General Research Fund from Research Grants Council of Hong Kong (Reference numbers: 464710, 475113, 14119219, 14119420, 14175617). Health and Medical Research Fund from Food and Health Bureau of Hong Kong (Reference numbers: 06171016, 07181266). Continuation project of Joint Research Fund for Overseas Chinese Scholars and Scholars in Hong Kong and Macao Young Scholars (Reference number: 31729002). National Natural Science Foundation of China (Reference numbers: 81971514, 82073950). Shenzhen Science and Technology Plan Project (Reference number: GJHZ20190822095605512, SGDX20201103095609027).
CONFLICT OF INTEREST STATEMENT
We have no competing interest to disclose.
IRB STATEMENT
This study was approved by the institutional review board (IRB no. 4-2013-0397) of Institute of Allergy, College of Medicine, Yonsei University for using the patient sera in ELISA experiments and the hospital ethics committee of The First Affiliated Hospital of Guangzhou Medical University (reference no. 2017018) for using the pooled patient sera in immunoblotting (western blotting) following the 2D gel electrophoresis.
Open Research
DATA AVAILABILITY STATEMENT
The genome and sequencing data of Tyrophagus putrescentiae are deposited in NCBI database under BioProject accession PRJNA706095. The in-silico-identified allergen sequences were uploaded to NCBI GenBank database under accessions OP558975–OP559059.




