Hippo pathway monomerizes STAT3 to regulate prostate cancer growth
Qingfeng Tang and Jing Fang contributed equally to this work.
Abstract
Prostate cancer ranks among the most commonly diagnosed malignancies for men and has become a non-negligible threat for public health. Interplay between inflammatory factors and cancer cells renders inflammatory tissue environment as a predisposing condition for cancer development. The Hippo pathway is a conserved signaling pathway across multiple species during evolution that regulates tissue homeostasis and organ development. Nevertheless, whether Hippo pathway regulates cancer-related inflammatory factors remains elusive. Here, we show that high cell density–mediated activation of the Hippo pathway blunts STAT3 activity in prostate cancer cells. Hippo pathway component MST2 kinase phosphorylates STAT3 at T622, which is located in the SH2 domain of STAT3. This phosphorylation blocks the SH2 domain in one STAT3 molecule to bind with the phosphorylated Y705 site in another STAT3 molecule, which further counteracts IL6-induced STAT3 dimerization and activation. Expression of a nonphosphorylatable STAT3 T622A mutant enhances STAT3 activity and IL6 expression at high cell density and promotes tumor growth in a mice xenograft model. Our findings demonstrate that STAT3 is a novel phosphorylation substrate for MST2 and thereby highlight a regulatory cascade underlying the crosstalk between inflammation and the Hippo pathway in prostate cancer cells.
Abbreviations
-
- ADT
-
- androgen deprivation therapy
-
- IL6
-
- interleukin 6
-
- JAK2
-
- Janus kinase 2
-
- LATS1/2
-
- large tumor suppressor 1/2
-
- MST1/2
-
- sterile 20-like kinase 1/2
-
- SH2
-
- Src homology 2
-
- STAT3
-
- signal transducer and activator of transcription 3
-
- TAZ
-
- tafazzin
-
- YAP1
-
- Yes-associated protein
1 INTRODUCTION
Prostate cancer ranks among the most commonly diagnosed malignancies for men and has become a non-negligible threat for public health with an estimated 350,000 deaths annually.1 Prostate malignant transformation is a multistep process, starting from prostatic intraepithelial neoplasia, then localized prostate cancer, which thereafter turns into local invasion of advanced prostate cancer with the degradation of basal cell layer and invasion of cancer cells through the basal layer, culminating metastatic prostate cancer.2 Androgen deprivation therapy (ADT) is currently the most common treatment for advanced and metastatic prostate cancer. Although most patients initially respond well to ADT, almost all cases will relapse, leading to castration-resistant prostate cancer.3 Therefore, a better understanding of the key factors involved in each step is important for improving the current diagnosis and treatment of prostate cancer.
The role of inflammation during cancer development was discovered by Rudolf Ludwig Carl Virchow in 1863, whose finding recognized the inflammatory tissue environment as a predisposing condition for carcinogenesis.4 The interleukin 6 (IL6)–signal transducer and activator of transcription 3 (STAT3) signaling axis is a major intrinsic pathway for cancer inflammation.5 IL6 exerts its biological effects by binding to IL6 receptor-α (IL6Rα) on the cell surface, which induces a conformational change to trigger the formation of a hexamer signaling complex, containing a gp130 (also known as IL6Rβ) homodimer and two IL6-IL6Rα heterodimers. These events lead to Janus kinase 2 (JAK2)-dependent activation of cytoplasmic STAT3, which facilitates its dimerization and translocation to the nucleus, functioning as a transcriptional factor to regulate the expression of a variety of inflammation- or cancer-related genes.6, 7 In cancer cells, abnormal IL6 expression and constitutive activation of STAT3 are closely related to an enhanced cancer cell proliferation and drug resistance.
The Hippo pathway is a conserved signaling pathway across multiple species during evolution that regulates tissue homeostasis and organ development.8, 9 Inactivation of the Hippo pathway is frequently observed in tumors due to a variety of mechanisms including mutations and dysregulation of upstream signaling factors.10 The core kinase cascade, composed by sterile 20-like kinase 1/2 (MST1/2) and large tumor suppressor 1/2 (LATS1/2), phosphorylates the major downstream effector proteins, Yes-associated protein (YAP1) and its paralog tafazzin (TAZ), leading to degradation of YAP1 and TAZ or sequestration of these two proteins in cytosol.11 Nevertheless, whether the Hippo pathway signals to impact STAT3 activation remains elusive.
In this study, we demonstrate that MST2 phosphorylates STAT3 at T622, which impairs STAT3 dimerization by altering its Src homology 2 (SH2) domain. MST2-mediated STAT3 phosphorylation blocks IL6-indcued activation of STAT3 and regulates prostate cancer growth.
2 MATERIALS AND METHODS
2.1 Materials
Rabbit polyclonal antibody recognizing phosphorylated STAT3 T622 was customized from Boer Biotechnology. To prepare antibody recognizing STAT3 pT622, rabbits were treated with peptide containing STAT3 pT622. Nonmodified peptide immobilized on an affinity column was used to remove the antibodies recognizing nonphosphorylated STAT3, and STAT3 pT622 peptide immobilized on an affinity column was used to associate with and isolate the antibodies. The eluted antibodies were then concentrated.
Antibodies recognizing TAZ (#83669), STAT3 (#9139), MST2 (#3952), MST1 (#14946), YAP1 (#12395), SAV1 (#13301), and STAT3-pY705 (#9145) were obtained from Cell Signaling Technology. Antibody simultaneously recognizing LATS1 and LATS2 (BS-4081R) was obtained from Bioss. Antibodies recognizing LATS1/2 pT1079/T1041 (ab111344), Ki67 (ab16667), YAP1 (ab205270), HA (ab9110), tubulin (ab7291), Flag (ab205606), GST (ab36415), YAP1 pS127 (ab76252), phospho-Thr (ab9337), JAK2 (ab108596), and recombinant IL-6 protein (ab9627) were purchased from Abcam. Anti-Flag agarose beads were obtained from Sigma. XMU-MP-1 was obtained from MedChemExpress. [γ-32P]-ATP was obtained from MP Biomedicals.
2.2 Cell culture
DU145 (RRID: CVCL_0105) and LNCaP (RRID: CVCL_0395) cells were purchased from ATCC. DU145 cell line was cultured with ATCC-formulated Eagle's minimum essential medium, and LNCaP cell line was cultured with ATCC-formulated RPMI-1640 medium. Medium was supplemented with 10% fetal bovine serum. All human cell lines have been authenticated using short tandem repeat (STR) profiling within the last 3 years. All experiments were performed with mycoplasma-free cells.
To generate stably shRNA-expressed cell lines, cells were expressed with shRNA construct and treated with puromycin to select the positive clones. To generate stably gene-expressed cell lines, a pCDH lentivector expression system (System Biosciences) was used according to the manufacturer's instructions. Briefly, the coding sequence of a gene was cloned into the pCDH lentiviral vector. Lentiviruses were generated by transfection of the aforementioned recombinant expression plasmid as well as pLP1, pLP2, pLP/VSVG vectors, which are required for virus packaging, in 293FT cells. The culture supernatants were collected 72 hours after transfection and centrifuged and filtered to remove cell debris. The infected cells were treated by hygromycin or G418 to select the positive clones.
2.3 Immunoprecipitation and immunoblot analysis
Immunoprecipitation and immunoblot analyses were performed following previous reports.12, 13 Cells were lysed with a buffer (0.1% SDS, 0.5 mM EDTA, 1% Triton X-100, 100 μM sodium pyrophosphate,150 mM NaCl, 100 μM PMSF, 100 μM leupeptin, 1 μM aprotinin, 1 mM dithiothreitol, 100 μM sodium orthovanadate, 50 mM Tris–HCl [pH 7.5], and 1 mM sodium fluoride). Cell lysates were centrifuged at 11,000 g, and the supernatants were incubated with indicated antibodies overnight at 4°C. Then, the agarose beads were applied to the mixture for incubation for another 3 hours. The protein-beads complexes were washed three times and then analyzed by immunoblot.
2.4 DNA constructs and mutagenesis
shRNAs were prepared using the following sequences: STAT3 shRNA, TAC CTA AGG CCA TGA ACT T (targeting noncoding region); MST1 shRNA, TAT AAT TCC TTC CAG GAC C; MST2 shRNA-1, TAT GTT CAA TCA TGG TCT G (this shRNA was used if the number of MST2 shRNA is not specified); MST2 shRNA-2, TTA ATT GCG ACA ACT TGA C; LATS2 shRNA, GCC ATG AAG ACC CTA AGG AAA; LATS1 shRNA, GAA GAT AAA GAC ACT AGG AAT; SAV1 shRNA, GTC TGA ATA ATA ATA TCG G; control shRNA, GCT TCT AAC ACC GGA GGT CTT.
Human YAP1, TAZ, STAT3, JAK2, and MST2 genes were cloned into pcDNA3.1/hygro(+)-Flag, pGEX-4 T-1, pcDNA3.1/hygro(+)-HA, or pCold I (His) vectors. QuikChange site-directed mutagenesis kit (Stratagene) was used to prepare the mutants. shRNA-resistant (r)MST2 was prepared by introducing three mutations (t1053a, a1056g, and t1059c) in the targeting site for shRNA-1.
2.5 Purification of recombinant proteins
GST-STAT3, Flag-STAT3, His-MST2, His-JAK2, and mutant proteins were expressed in BL21(DE3) bacteria as previously described.14 Bacteria cells were cultured in Lysogeny Broth medium, and expression of these proteins was induced by IPTG for 16 hours at 30°C or 16°C, followed by lysis via sonication.
To purify the His-MST2 and His-JAK2, the lysed bacterial samples were transferred to a Ni-NTA column (GE Healthcare Life Sciences). The column was flushed with 20 mM imidazole, and the protein was eluted with 250 mM imidazole.
To purify Flag-STAT3, the lysed bacterial samples were transferred to a column containing Anti-Flag M2 agarose affinity gel. The column was flushed with PBS and the protein was eluted with 100 μg/mL Flag peptide.
To purify GST-STAT3, the lysed bacterial samples were transferred to a GSTrap HP column (GE Healthcare Life Sciences). The column was flushed with PBS, and the protein was eluted with 10 mM reduced glutathione.
To remove contaminated proteins, the eluted samples were separated through a HiPrep 16/60 Sephacryl S-200 HR gel filtration column (GE Healthcare Life Sciences).
2.6 RT-PCR
Total mRNA was isolated from cells by using the RNeasy Micro Kit (Qiagen). QuantiTect Reverse Transcription Kit (Qiagen) was then used to produce cDNA, and RT-PCR was performed using the following primers: IL6-F: ACT CAC CTC TTC AGA ACG AAT TG; IL6-R: GCA TCT AGA TTC TTT GCC TTT TTC TG; Actin-F, CAT GTA CGT TGC TAT CCA GGC; Actin-R, CTC CTT AAT GTC ACG CAC GAT.
2.7 In vitro kinase assay
In vitro kinase assay for MST2 or JAK2 was performed according to a previous method.15 A total of 20 ng purified kinase protein was mixed with 50 ng purified STAT3 protein in the reaction buffer containing 50 mM potassium acetate, 30 mM HEPES, 500 μM ATP, and 5 mM MgCl2 for 30 minutes at 30°C. If autoradiography was used as the detection method, 10 μCi [γ-32P]ATP was included in the reaction system. The reaction was terminated by boiling in sample buffer, and the protein samples were separated by SDS-PAGE. The phosphorylation was detected by immunoblotting with indicated antibodies recognizing phosphor-Thr or STAT3-pT622, or by autoradiography.
2.8 Size-exclusion chromatography analysis
Size-exclusion chromatography was performed using a HiPrep 16/60 Sephacryl S-300 HR column (GE), following a previous report.16 A total of 20 μg protein samples were fractionated at a rate of 0.5 mL/minute. The eluted samples were collected for 1 mL for each soluble fraction and subjected to immunoblotting assay. A size-exclusion chromatography calibration marker kit (Sigma) was used to establish the standard according to the manufacturer's instructions.
2.9 STAT3-DNA binding assay
STAT3-DNA binding assay was measured by using a STAT3 transcription factor assay kit (Colorimetric) obtained from Abcam (ab207229), following the manufacturer's instructions.
2.10 STAT3 transcription activity assay
STAT3 transcription activity assay was performed by using a STAT3 reporter kit obtained from BPS Bioscience (#79730), following the manufacturer's instructions.
2.11 BrdU incorporation assay
BrdU incorporation assay was performed by using a BrdU cell proliferation ELISA kit (colorimetric) obtained from Abcam (ab126556), following the manufacturer's instructions.
2.12 Mice tumor xenograft model
A total of 5 × 106 cells were subcutaneously introduced into 6-week-old male BALB/c nude mice. Mice were sacrificed 24 days after implantation, and the tumor size was determined by the formula: 0.5 × length × width2. The use of animals was approved by the Institutional Review Board of Chengdu Medical College. The animals were treated in accordance with relevant institutional and national guidelines and regulations.
2.13 Immunohistochemical staining of clinical samples
VECTASTAIN ABC kit (Vector Laboratories) was used to perform the immunohistochemical staining, which was then quantified by two pathologists following a previously reported scoring method.17 Proportion scores were defined as: 0, no tumor cells were positive stained; 1, 1%-10%; 2, 11%-30%; 3, 31%-70%; and 4, 71%-100%. Intensity score was defined as: 0, negative; 1, weak; 2, moderate; 3, strong; 4, very strong. We then multiplied the proportion and intensity scores to obtain a total score. A total of 55 prostate cancer tissues were obtained with informed consent from all subjects. The use of human prostate cancer specimens was approved by the Institutional Review Board at Xindu District People's Hospital of Chengdu.
2.14 Quantification and statistical analysis
Statistical analyses were performed using two-sided Student's t test for comparison between two groups or ANOVA test for multiple groups. All data represent the mean ± SD of at least three independent experiments unless otherwise specified. Differences in means were considered statistically significant at p < 0.05.
3 RESULTS
3.1 MST2 impairs IL6-induced STAT3 activation at high cell density
To determine the impact of the Hippo pathway on STAT3 activation in prostate cancer cells, DU145 and LNCaP cells were harvested at high (~80%) or low (~10%) cell confluence. As expected, an apparent inactivation of the Hippo pathway, shown by the reduced phosphorylation of LATS1/2 and increased expression of YAP1 and TAZ, was observed in those cells at low confluence (Figure 1A). STAT3 activity assay, using a luciferase reporter gene under the transcriptional control of a minimal STAT3 response element, revealed a modest increase in STAT3 activity at low cell density, which was in line with a previous report that YAP and TAZ are coactivators of STAT3.18 Strikingly, treatment with IL6 recombinant protein largely boosted STAT3 activity at low cell density; in contrast, only minor changes were found in STAT3 activity in the cells cultured at high cell density (Figure 1A). Further, incubation with a Hippo pathway inhibitor, XMU-MP-1, which abolished LATS1/2 phosphorylation and restored YAP1/TAZ expression at high confluence, markedly enhanced IL6-elicited STAT3 activity in DU145 and LNCaP cells (Figure 1B). The altered level of IL6-induced STAT3 activity at different cell densities was not likely due to the gp130/JAK2 signaling complex, as almost equal STAT3 Try(Y)705 phosphorylation was detected (Figure 1A,B).
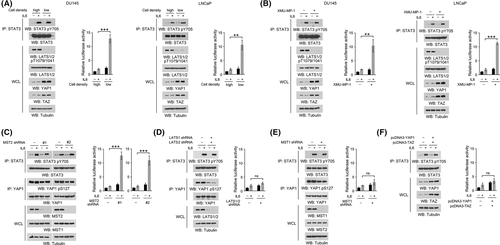
Next, we moved to identify the Hippo pathway component that regulated STAT3 activity. Depletion of MST2 by two distinct shRNAs increased STAT3 activity fivefold to sevenfold in DU145 cells at high cell density in response to IL6 treatment (Figure 1C). Nevertheless, no obvious change was observed upon knockdown of LATS1/2 or MST1 or overexpression of YAP1/TAZ (Figure 1D–F). These results suggest that MST2 impairs IL6-induced STAT3 activation at high cell density.
3.2 MST2 phosphorylates STAT3 at T622
As MST2 is a protein kinase, of our particular interest, we examined whether STAT3 is a direct substrate for MST2. We mixed the bacterially purified GST-STAT3 protein with the purified active WT His-MST2 protein in the presence of radioactive [γ-32P]-ATP for an in vitro kinase assay. Notably, an apparent autoradiographic signal was detected in STAT3 protein after incubation with purified WT MST2 protein (Figure 2A). In contrast, a kinase-dead MST2 K56R mutant,19 which had only one residue replaced compared to its WT counterpart, did not cause a similar autoradiographic signal (Figure 2A). As both the WT and mutant MST2 proteins were purified through the same protocol, the autoradiographic signal understandably resulted from the phosphorylation mediated by MST2, rather than the contaminated unknown kinases in the purified MST2 protein.
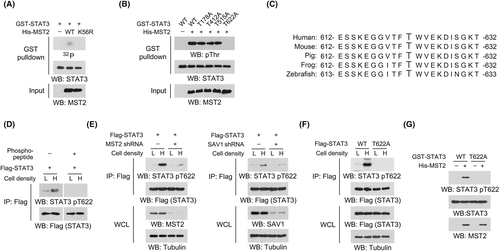
Analyses of STAT3 protein sequence revealed four putative threonine phosphorylation sites, according to a previously reported MST2 phosphorylation motif.20, 21 Replacement of these threonine into alanine revealed that only T622A mutation abrogated MST2-meidated phosphorylation (Figure 2B). Alignment analyses showed that STAT3 T622 site was evolutionally conserved across multiple species (Figure 2C). We therefore prepared an antibody recognizing phosphorylated STAT3 T622 and detected a markedly enhanced STAT3 T622 phosphorylation at high cell density (Figure 2D). Detection of this phosphorylation could be abolished by incubation with a STAT3 pT622 blocking peptide (Figure 2D), suggesting a good specificity of this antibody. As expected, the use of MST2 shRNA or STAT3 T622A mutant substantially obliterated STAT3 T622 phosphorylation induced by high cell density in DU145 cells or by purified His-MST2 protein in an in vitro kinase assay (Figure 2E–G). Consistently, knockdown of SAV1, a critical factor for the activation of both MST1 and MST2,22 also impeded STAT3 T622 phosphorylation (Figure 2E). These results suggest that MST2 phosphorylates STAT3 at T622.
3.3 MST2-dependent STAT3 T622 phosphorylation impairs STAT3 dimerization and activation
STAT3 uses its SH2 domain to bind with the phosphorylated Y705 site of another STAT3 molecule, which is the essential step for STAT3 activation.6 Analyses of the sequence and structure (protein data base (PDB) code: 6TLC) of human STAT3 revealed that T622 was located in the peptide-binding cleft of the SH2 domain (Figure 3A,B). We thus wondered whether MST2-dependent STAT3 T622 phosphorylation could affect STAT3 intermolecular interaction. Indeed, shift from high to low cell density markedly increased the IL6-induced association between Flag-STAT3 and HA-STAT3, accompanied by an inhibited Hippo pathway (Figure 3C). Either knockdown of MST2 or expression of STAT3 T622A mutant also largely enhanced this interaction at high cell density (Figure 3D,E). To evaluate the direct interaction between STAT3 molecules, bacterially purified WT GST-STAT3 and WT Flag-STAT3 proteins were sequentially subjected to His-MST2 protein– and His-JAK2 protein–dependent in vitro kinase assay. Though MST2-mediated T622 phosphorylation had no obvious impact on JAK2-mediated STAT3 Y705 phosphorylation, it largely blocked JAK2-induced interaction between WT GST-STAT3 and WT Flag-STAT3 proteins (Figure 3F). In contrast, the interaction between nonphosphorylatable STAT3 T622A mutant proteins was nearly intact (Figure 3F). In addition, the aforementioned manipulations did not change the low binding affinity of STAT3 proteins in the absence of IL6 or JAK2 treatment (Figure 3C–F), suggesting that STAT3 Y705 phosphorylation was indispensable for STAT3 intermolecular interaction.
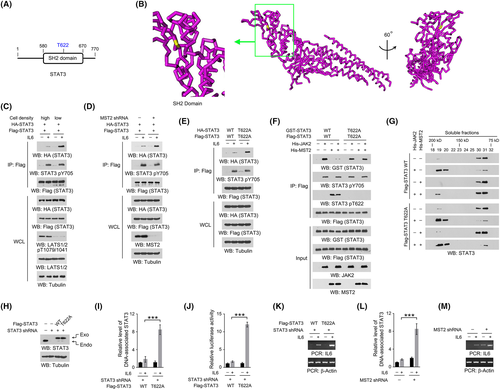
To further determine the oligomerization status of STAT3 protein, the bacterially purified Flag-STAT3 proteins derived from the sequential His-MST2 protein– and His-JAK2 protein–dependent in vitro kinase assay were subjected to size exclusion chromatography assay. According to the molecular weight calibration standard, untreated WT STAT3 protein was mainly eluted in soluble fractions 30-31, which contained proteins with a molecular weight around 80-90 kDa. JAK2-induced phosphorylation enriched the STAT3 protein in the fractions 19-20, which contained proteins with a molecular weight around 170-190 kDa. Though MST2-induced phosphorylation did not change the monomer status of the unmodified STAT3 protein, it kept STAT3 protein mainly as monomer even under JAK2-induced phosphorylation. Notably, STAT3 T622A mutant protein was responsive to JAK2-induced dimerization regardless of MST2-mediated effects (Figure 3G), suggesting that MST2-dependent STAT3 T622 phosphorylation impairs STAT3 dimerization.
Further, we stably depleted endogenous STAT3 in DU145 cells and exogenously expressed equal amounts of WT or T622A-mutated STAT3 proteins (Figure 3H). A DNA pulldown assay showed that, at high cell density, a double-stranded DNA containing the STAT3 binding consensus (5-TTCCCGGAA-3) interacted with much more T622A mutant STAT3 protein than the WT control protein under IL6 treatment (Figure 3I). Consistently, T622A mutation markedly elevated the STAT3 transcriptional activity and the level of IL6 mRNA (Figure 3J,K). Similar results were obtained by knockdown of MST2 (Figure 3L,M). These results suggest that MST2-dependent STAT3 T622 phosphorylation counteracts STAT3 activation.
3.4 MST2-dependent STAT3 T622 phosphorylation impairs prostate cancer growth
We next determined the impact of STAT3 T622 phosphorylation on the proliferation of prostate cancer cells. BrdU incorporation assay revealed that, when cultured at high density, IL6 treatment induced a more pronounced proliferation in two different DU145 clones with STAT3 T622A mutant, compared with the clone with WT STAT3. However, no apparent differences were found between the IL6-untreated groups (Figure S1A). Similar effects were also found when endogenous WT MST2 was replaced by a kinase-dead MST2 K56R mutant (Figure S1B). To extend our finding, we then established a mouse xenograft model by subcutaneous injection of DU145 (Figure 3H) or LNCaP (Figure S1C) cells with WT or mutant STAT3. As expected, expression of STAT3 T622A mutant palpably enhanced tumor growth with a markedly increased tumor volume (Figure 4A,B and S1D). Notably, knockdown of IL6 substantially attenuated tumor formation by STAT3 T622A mutated cells (Figure S1D,E), suggesting an important role of IL6 in promoting tumor cell growth. With the valid STAT3 pT622 antibody (Figure 4C), we found that T622A mutation abolished STAT3 T622 phosphorylation, and substantially increased the expression of IL6 and proliferative cell marker Ki-67 (Figure 4D). These results suggest that MST2-dependent STAT3 T622 phosphorylation impairs prostate cancer growth.
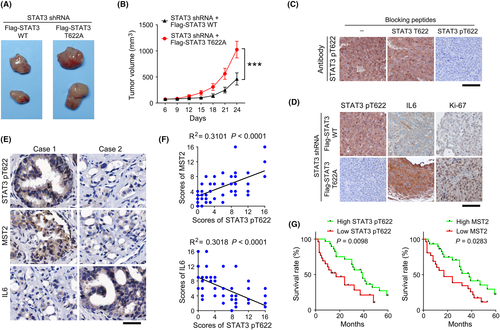
3.5 STAT3 T622 phosphorylation correlates with clinical aggressiveness of prostate cancer
We next performed IHC staining using clinical samples derived from prostate cancer surgery and found that STAT3 T622 phosphorylation was positively correlated with MST2 expression but negatively correlated with IL6 expression (Figure 4E). The linear regression analyses showed that these correlations were statistically significant (Figure 4F). Additionally, those patients who had high levels of STAT3 T622 phosphorylation or MST2 expression showed a longer survival time (Figure 4G). These results suggest a close relationship between STAT3 T622 phosphorylation and clinical aggressiveness of prostate cancer.
4 DISCUSSION
The Hippo pathway, which exerts a critical role in modulating mechano-transduction under diverse cell densities or cell-environment interactions, is frequently reported to be downregulated in a broad range of human cancers.23 MST1/2 and LATS1/2 kinases constitute the core phosphorylation cascade of the Hippo pathway, which eventually tunes YAP1/TAZ to regulate tumor growth.24 Hitherto, accumulating new phosphorylation substrates, functioning in diverse biological processes, have been discovered for Hippo pathway kinases, which largely expand the existing knowledge on the roles of the Hippo pathway in the interconnection with other signaling pathways.25 Recently, our lab has reported that LATS1/2 phosphorylates and inhibits phosphoribosyl pyrophosphate synthetase, the rate-limiting enzyme for nucleotide synthesis, thereby impeding DNA and RNA synthesis in response to softened extracellular environment.9 In this study, we find that STAT3 is a new phosphorylation substrate of MST2. By in vitro kinase assay using [γ-32P]-ATP, we show that MST2 recombinant protein, but not MST1, LATS1, LATS2, or kinase-dead MST2 K56R mutant protein, introduced an autoradiographic signal on STAT3 protein. We further identified STAT3 T622 as the phosphorylation site, as only the mutation of this site into a nonphosphorylatable alanine abolished MST2-mediated phosphorylation.
YAP1 and TAZ are well established downstream targets of Hippo kinases. When the Hippo pathway is activated, LATS1/2 directedly phosphorylate YAP1/TAZ at multiple sites, which reinforces the cytosolic localization and proteolytic degradation of YAP1/TAZ. In contrast, without LATS1/2-mediated phosphorylation, active YAP/TAZ accumulate in the nucleus and bind with TEA domain family member (TEAD) transcription factors, so they elicit the transcription of a large body of genes functioning in cell growth and differentiation.26 In line with previous reports, we found that simultaneous knockdown of LATS1/2 largely reduced YAP1 phosphorylation in the cells cultured in high density, while knockdown of MST1 or MST2 alone only resulted in a slight or modest decrease, probably because one MST could substantially compensate for the other MST in the Hippo phosphorylation cascades. Further, overexpression of YAP1 and TAZ did not induce a comparable increase in STAT3 activity, as did MST2 knockdown, suggesting that YAP1/TAZ may have minor effects on the MST2-mediated regulation of STAT3.
STAT3 has been evidenced to be involved in diverse cellular processes, such as proliferation, migration, and responses to stresses.27 Notably, hyperactivation of STAT3 is frequently found in various human tumors and shows a potential value in predicting patient outcome. Given the important roles of STAT3 in tumor development, it is not surprising that STAT3-mediated signaling transduction and the regulatory mechanisms of STAT3 activation are widely regarded as potential targets for tumor treatment.28 In this study, we report an important molecular mechanism that modulate STAT3 activation, a central regulator in cancer-related inflammation. STAT3 strongly binds to DNA when it is in the dimeric form. STAT3 dimerization is carried out by its SH2 domain, as SH2 domain from one STAT3 monomer associates with the phosphorylated Y705 located in the transactivation domain of another STAT3 monomer.29 We demonstrate that MST2-mediated phosphorylation at STAT3 T622 dampened the STAT3 dimerization, thereby inhibiting STAT3 activation. Further, prostate cancer cells harboring STAT3 T622A grew much faster than the WT cells, and patients with a high level of STAT3 T622 phosphorylation showed a longer average survival time. The current data add to the knowledge of the regulatory mechanism responsible for STAT3 activation and may assist the development of novel STAT3 antagonist for cancer therapy.
Hyperactivation of STAT3 plays a critical role in malignant initiation, tumor progression, and metastatic dissemination.30 In this study, our findings illustrate a new regulatory mechanism implicated in cancer-related inflammation. Cell density signals to govern the activation of oncogene STAT3 by modulating its SH2 domain in an MST2-dependent manner. Disruption of this Hippo pathway–guided inflammatory cascade benefits prostate cancer cell growth.
ACKNOWLEDGMENTS
This work was supported by Bethune Public Welfare Fund ZHYX201809004 (Q.T.) and a fund of the Chengdu Municipal Health Commission 2021141 (Y.H.).
DISCLOSURE
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
ETHICS STATEMENT
All clinical tissues were obtained with informed consent. The use of human prostate cancer specimens was approved by the Institutional Review Board at Xindu District People's Hospital of Chengdu. The use of animals was approved by the Institutional Review Board of Chengdu Medical College. The animals were treated in accordance with relevant institutional and national guidelines and regulations.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




