Development of a potent small-molecule degrader against oncogenic BRAFV600E protein that evades paradoxical MAPK activation
Funding Information
This work was supported in part by grants from the Japan Society for the Promotion of Science (KAKENHI grants JP18K06567 and JP21K06490 to N.O. and JP18H05502 and JP21H02777 to M.N.), the Japan Agency for Medical Research and Development (AMED grants JP19cm0106136, JP21ak0101073, and JP21mk0101197 to N.O. and JP20ak0101073 to M.N.), and the Princess Takamatsu Cancer Research Fund (to M.N.).
Abstract
BRAF mutations are frequently observed in melanoma and hairy-cell leukemia. Currently approved rapidly accelerated fibrosarcoma (RAF) kinase inhibitors targeting oncogenic BRAF V600 mutations have shown remarkable efficacy in the clinic, but their therapeutic benefits are occasionally hampered by acquired resistance due to RAF dimerization–dependent reactivation of the downstream MAPK pathway, which is known as paradoxical activation. There is also a concern that paradoxical activation of the MAPK pathway may trigger secondary cancer progression. In this study, we developed chimeric compounds, proteolysis targeting chimeras (PROTACs), that target BRAFV600E protein for degradation. CRBN(BRAF)-24, the most effective chimera, potently degraded BRAFV600E in a ubiquitin-proteasome system (UPS)-dependent manner and inhibited the proliferation of BRAFV600E-driven cancer cells. In BRAF wild-type cells, CRBN(BRAF)-24 induced neither BRAFWT degradation nor paradoxical activation of the MAPK pathway. Biochemical analysis revealed that CRBN(BRAF)-24 showed more potent and sustained suppression of MAPK signaling than a BRAFV600E inhibitor, PLX-8394, in BRAFV600E-driven cancer cells. Targeted degradation of BRAFV600E by CRBN(BRAF)-24 could be a promising strategy to evade paradoxical activation of the RAF-MAPK pathway.
Abbreviations
-
- CRBN
-
- cereblon
-
- IAP
-
- inhibitor of apoptosis protein
-
- MDM2
-
- murine double minute 2
-
- PROTAC
-
- proteolysis targeting chimera
-
- RAF
-
- rapidly accelerated fibrosarcoma
-
- RTK
-
- receptor tyrosine kinase
-
- SNIPER
-
- specific and nongenetic inhibitor of apoptosis protein (IAP)-dependent protein eraser
-
- UPS
-
- ubiquitin-proteasome system
-
- VHL
-
- von Hippel-Lindau
-
- WT
-
- wild-type
1 INTRODUCTION
RAF family kinases (ARAF, BRAF, and CRAF in humans) play a major role in cell proliferation, growth, differentiation, and survival.1-3 When receptor tyrosine kinases (RTKs) are activated by binding of extracellular growth factors, the small GTPase RASs recruit RAFs to the plasma membrane, promoting their dimerization and activation.4 Activated RAF phosphorylates and activates the downstream MEK and ERK kinases, thus transmitting growth signals through a kinase-activation cascade (known as the RAS-RAF-MEK-ERK pathway or MAPK pathway).
Among the genes encoding the three RAF proteins, BRAF is most prone to mutation in cancer cells. BRAF mutations are observed in melanomas, colorectal cancers, non–small cell lung cancers, and in almost all hairy-cell leukemia.5-8 Most of these mutations are missense mutations found in the kinase domain and affect the biochemical characteristics of BRAF kinase.9-11 Dysregulation of the MAPK signaling pathway by BRAF mutations is a potent driver of cancer development and progression.1-3
BRAF mutations are grouped into three classes. Class 1 BRAF mutations, sharing more than 90% of total BRAF alterations, include only BRAF V600 mutations (primarily BRAFV600E mutations).12, 13 These mutations produce constitutively active BRAF kinases capable of signaling as monomers, and their robust kinase activity results in high phosphorylation and activation levels of downstream ERK. Class 2 BRAF mutants, such as K601E, L597Q, and G469A, signal as constitutively active mutant dimers. The ability of class 1 and class 2 mutants to activate MAPK signaling is independent of upstream RAS activity, and highly activated ERK drives feedback inhibition of RAS activation.14-16 In contrast, class 3 BRAF mutants, such as G466V and D594N, have little or no kinase activity, and increase ERK signaling through heterodimers formed with wild-type RAFs.17 The class 3 BRAF mutants bind more tightly to RAS than wild-type BRAF and enhance the binding and activation of wild-type CRAF, the predominant partner for heterodimerization. Therefore, activation of ERK in tumors with the class 3 mutants requires RAS activation.
Selective RAF inhibitors, such as vemurafenib and dabrafenib, have shown remarkable clinical utility against highly active BRAFV600E mutant cancers.18 However, as with many kinase inhibitors resistance develops, and patients become insensitive during long-term treatment.19 In addition, these drugs paradoxically activate the MAPK pathway in BRAF wild-type cells, especially those with oncogenic RAS mutations or with elevated upstream receptor signaling.9-11 The paradoxical activation of the MAPK pathway raises concerns that RAF inhibitors may promote cell proliferation and facilitate the progression of secondary RAS-driven cancers.13, 20-23 Therefore, there is a need for alternative therapeutic strategies targeting BRAF that can evade paradoxical activation of MAPK signaling.
Targeted protein degradation using bifunctional small molecules such as proteolysis-targeting chimeras (PROTACs) and specific and non-genetic inhibitor of apoptosis protein (IAP)-dependent protein erasers (SNIPERs) is an emerging modality in drug discovery.24, 25 These molecules have a chimeric structure in which a ligand that binds to a target protein and another ligand that binds to an intracellular E3 ubiquitin ligase (e.g., CRBN, VHL, IAP, MDM2) are conjugated by a linker.26-31 The chimeras cross-link target proteins of interest and E3 ligases and promote ubiquitination and degradation of the target proteins by the 26S proteasome. By substituting target ligands, numerous PROTACs and SNIPERs have been developed that degrade pathogenic proteins involved in various diseases.26-33 In this study, we developed new chimeric molecules that degrade BRAF mutant proteins. The resulting molecule CRBN(BRAF)-24 shows potent activity to degrade BRAFV600E and does not induce paradoxical activation of the MAPK pathway in BRAFWT cells.
2 MATERIALS AND METHODS
2.1 Design and synthesis of PROTAC compounds
We designed PROTAC compounds in which a BRAF inhibitor is connected to a ligand for an E3 ligase via a linker. The chemical synthesis, schemes, and physicochemical data for compounds are provided in the Doc. S1 and Scheme S1-S23.
2.2 Reagents
Tissue culture plastics were purchased from Greiner Bio-One. RPMI 1640 medium, DMEM, Eagle's minimum essential medium, and kanamycin were from Merck. McCoy's 5a medium and Leibovitz's L-15 medium were from Thermo Fisher Scientific. Benzyloxycarbonyl-L-leucyl-L-leucyl-L-leucinal (MG132) was from Peptide Institute. MLN7243 and MLN4924 were from Active Biochem.
2.3 Cell culture
Human melanoma A375 cells, N-RAS mutant-A375 isogenic cells, and K-RAS mutant-A375 isogenic cells were maintained in DMEM containing 10% FBS and 100 μg ml−1 kanamycin. Human colon carcinoma COLO205, RKO, and NCI-H508 cells; human lung carcinoma NCI-H23, NCI-H1395, NCI-H2087, NCI-H1755, and NCI-H1666 cells; human melanoma SK-MEL-30 cells; human ovarian carcinoma OVCAR8 cells; and human B cell leukemia JVM-3 cells were maintained in RPMI 1640 medium containing 10% FBS (5% FBS for NCI-H1666) and 100 μg ml−1 kanamycin. Human melanoma SK-MEL-28 cells were maintained in Eagle's minimum essential medium containing 10% FBS and 100 μg ml−1 kanamycin. Human colon carcinoma HCT116 cells were maintained in McCoy's 5a medium containing 10% FBS and 100 μg ml−1 kanamycin. Human breast carcinoma MDA-MB-231 cells were maintained in Leibovitz's L-15 medium containing 10% FBS and 100 μg ml−1 kanamycin. SK-MEL-30 and JVM-3 cell lines were purchased from DSMZ. OVCAR8 cell line was described elsewhere.34 The other cell lines were purchased from ATCC. These cells were treated with various concentrations of compounds for the indicated periods of time.
2.4 Western blotting
Cells were lysed with SDS lysis buffer (0.1 M Tris-HCl at pH 8.0, 10% glycerol, 1% SDS) and immediately boiled for 10 minutes to obtain clear lysates. Protein concentrations were measured using the BCA method (Pierce). Lysates containing equal amounts of proteins were separated by SDS-PAGE and transferred to PVDF membranes (Merck) for Western blot analysis using the appropriate antibodies. Immunoreactive proteins were visualized using the Immobilon Western chemiluminescent HRP substrate (Merck) or Clarity Western ECL substrate (Bio-Rad); light emission intensity was quantified using an LAS-3000 lumino-image analyzer equipped with Image Gauge v2.3 software. The antibodies used in this study were: anti-BRAF antibody (14814; Cell Signaling Technology), anti-CRAF antibody (53745; Cell Signaling Technology), anti-MEK1/2 antibody (8727; Cell Signaling Technology), anti-phospho-MEK1/2 (Ser217/221) antibody (9121; Cell Signaling Technology), anti-p44/42 MAPK (ERK1/2) antibody (4695; Cell Signaling Technology), anti-phospho-p44/42 MAPK (ERK1/2) (Thr202/Tyr204) antibody (4377; Cell Signaling Technology), anti-RAS antibody (8955; Cell Signaling Technology), anti-CRBN antibody (71810; Cell Signaling Technology), and anti-β-actin antibody (A5316; Merck).
2.5 siRNA transfection
A375 cells were transiently transfected with an equal mixture of three CRBN-specific stealth short interfering RNAs (stealth siRNAs) (HSS121807, HSS121808, HSS121809; Life Technologies) or a negative control stealth siRNA (12935300; Life Technologies) using Lipofectamine RNAi MAX reagent (Life Technologies) according to the protocols provided by the manufacturer.
2.6 Cell viability assay
Cell viability was determined using water-soluble tetrazolium WST-8 (4-[3-(2-methoxy-4-nitrophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzenedisulfonate) reagent in a spectrophotometric assay according to the manufacturer's instructions (Dojindo). Cells treated with compounds were incubated with WST-8 reagent for 0.5 hour at 37°C in a humidified atmosphere of 5% CO2. The absorbance of the medium at 450 nm or 490 nm was measured using an EnVision Multilabel Plate Reader (PerkinElmer).
3 Results
3.1 CRBR(BRAF)-24 induces efficient and potent degradation of BRAFV600E
To target BRAF mutant proteins for degradation, we designed and synthesized several chimeric molecules that combine various BRAF inhibitors with four E3 ligase ligands (LCL161 for IAP,28 VH032 for VHL,27 pomalidomide for CRBN,35 and RG7388 for MDM230). After evaluating the ability of the chimeric compounds to degrade BRAFV600E mutant proteins in A375 cells (a cell line with homozygous BRAFV600E), we identified an active compound CRBN(BRAF)-1 composed of a clinical BRAF inhibitor PLX839423, 36, 37 and pomalidomide (Figure 1). As shown in Figure 2A, CRBN(BRAF)-1 effectively reduced BRAF protein levels after 24 hours of treatment. CRBN(BRAF)-2, in which the same ligands are conjugated with a different linker, showed attenuated activity to reduce BRAFV600E (Figure 1 and Figure 2A). Chimeric compounds recruiting other E3 ligases (IAP[BRAF]-7, VHL[BRAF]-2, and MDM2[BRAF]-2) that replaced the pomalidomide of CRBN(BRAF)-1 with LCL161, VH032, and RG7388, respectively, did not reduce BRAF levels (Figure S1).

We further developed a series of chimeric compounds (CRBN[BRAF]-23, CRBN[BRAF]-24, CRBN[BRAF]-12, and CRBN[BRAF]-8) with improved degradation activity by modifying CRBN(BRAF)-1 with linkers of different lengths and structures (Figure 1). CRBN(BRAF)-24, one of the most potent compounds, was selected as a representative on the basis of its ability to reduce BRAF protein levels in A375 cells (Figure 2B,C). CRBN(BRAF)-24 showed a DC50 (the concentration at which 50% of maximum degradation was observed) value of 6.8 nM and DMAX (the percentage of maximum degradation observed) of approximately 80% in A375 cells at 24 hours after treatment (Figure 2B,C). The reduction in BRAF protein was maintained for at least 72 hours (Figure 2D). In SK-MEL-28 cells (another cell line with homozygous BRAFV600E), CRBN(BRAF)-24 also showed effective BRAF-reducing activity and inhibited the downstream pathway, MEK, as shown by suppressed ERK phosphorylation (Figure 2E). We further synthesized an inactive degrader, CRBN(BRAF)-29, by methylating the nitrogen of the glutarimide ring in the pomalidomide moiety of CRBN(BRAF)-24, which abolished the recruitment of CRBN35 (Figure 1). CRBN(BRAF)-29 did not decrease BRAF protein levels, suggesting that the binding to CRBN with a pomalidomide moiety is necessary for the BRAFV600E reduction by CRBN(BRAF)-24 (Figure 2E). In addition, the activity of CRBN(BRAF)-29 to inhibit the downstream kinase signaling was approximately 10-fold weaker than that of CRBN(BRAF)-24, suggesting that the BRAF degradation activity of CRBN(BRAF)-24 contributes to better inhibition of downstream kinase signaling. A moderate hook effect was occasionally observed in the CRBN(BRAF)-24–induced BRAF reduction at higher concentrations (Figure 2B and D), probably because excess amounts of bivalent compounds inhibited the formation of the ternary complex composed of target (BRAF), degrader (CRBN[BRAF]-24), and E3 ligase (CRBN).24, 27
3.2 CRBN(BRAF)-24 degrades BRAFV600E via UPS
To determine whether CRBN(BRAF)-24 reduces the protein level of BRAFV600E by the expected molecular mechanism, we first investigated the effect of UPS inhibitors. When A375 cells were treated with a proteasome inhibitor, MG132, or a ubiquitin-activating enzyme inhibitor, MLN7243, together with CRBN(BRAF)-24, the CRBN(BRAF)-24–induced reduction in BRAF levels was completely suppressed (Figure 3A). Similarly, the effect of CRBN(BRAF)-24 was also inhibited by MLN4924, a NEDD8-activating enzyme inhibitor that inhibits the activity of the Cullin-RING E3 ligases (CRLs), including CRL4CRBN. These results suggest that CRBN(BRAF)-24 degrades BRAFV600E via UPS using the CRL-type E3 ligases. It was also confirmed that treatment with a mixture of the BRAF ligand (PLX8394-L) and the CRBN ligand (pomalidomide) did not induce BRAF degradation (Figure 1; Figure 3B). In addition, cotreatment with an excess amount of PLX8394-L (target ligand competition) or pomalidomide (E3 ligand competition) inhibited the BRAF degradation by CRBN(BRAF)-24 (Figure 3B). Silencing the expression of CRBN by RNA interference (RNAi) inhibited the degradation of BRAF by CRBN(BRAF)-24 (Figure 3C). These results collectively indicate that CRL4CRBN is required for BRAFV600E degradation by CRBN(BRAF)-24.

3.3 Neither BRAF degradation nor paradoxical activation of MAPK signaling is induced by CRBR(BRAF)-24 in BRAFWT cells
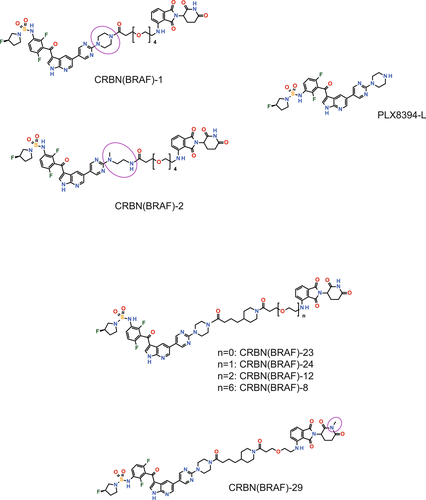
PLX8394, the BRAF ligand used for CRBN(BRAF)-24, has been shown to bind to BRAFV600E, BRAFWT, and CRAF, although the binding is approximately three- and sixfold more selective for BRAFV600E than for BRAFWT and CRAF, respectively.23 Therefore, we next examined if CRBN(BRAF)-24 can degrade BRAFWT and CRAF proteins. In cell lines that do not bear the BRAF mutant alleles (HCT116, NCI-H23, SK-MEL-30, and OVCAR8), CRBN(BRAF)-24 had little effect on BRAFWT and CRAF protein levels (Figure 4B), strongly suggesting that CRBN(BRAF)-24 did not induce the degradation of BRAFWT and CRAF proteins. In cell lines bearing BRAFV600E alleles, CRBN(BRAF)-24 showed marginal (A375, COLO-205, and RKO) or moderate (SK-MEL-28) activity to reduce CRAF protein (Figure 4A). However, the CRAF reduction mechanism in these cells seems to be different from the BRAFV600E degradation because depletion of CRBN did not affect the CRAF reduction (Figure S2A), and CRAF protein levels were also reduced by PLX8394 (Figure S2B). Importantly, CRBN(BRAF)-24 did not activate MAPK signaling in any BRAFWT cells regardless of the presence of oncogenic RAS (Figure 4B), indicating that CRBN(BRAF)-24 does not induce paradoxical activation even in cells in which BRAF is constantly activated by the oncogenic RAS mutants.
3.4 CRBN(BRAF)-24 selectively induces growth inhibition of cell lines expressing BRAFV600E
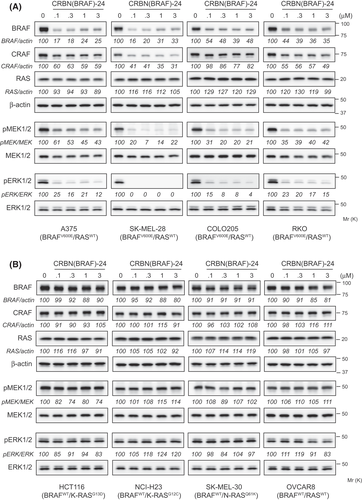
Next, we investigated the effect of CRBN(BRAF)-24 on cancer cell proliferation and compared its efficacy with that of CRBN(BRAF)-29 and PLX8394. Consistent with the effective inhibition of the downstream MAPK pathway (Figure 4A), CRBN(BRAF)-24 strongly inhibited proliferation in three of the four BRAFV600E cell lines (A375, SK-MEL-28, and COLO205) at 10 nM and higher (Figure 5A). In these cell lines, CRBN(BRAF)-24 demonstrated better antiproliferative activity than CRBN(BRAF)-29 and PLX8394. In RKO cells, CRBN(BRAF)-24 showed a weak activity to inhibit proliferation. In contrast, none of the drugs showed antiproliferative activity against BRAFWT cell lines (Figure 5B). These results indicate that CRBN(BRAF)-24 selectively inhibits proliferation of cancer cells harboring BRAFV600E mutation, as does PLX8394.
3.5 CRBN(BRAF)-24 exerts a potent and sustained inhibitory effect by degrading BRAFV600E
Because CRBN(BRAF)-24 uses a BRAF inhibitor as a warhead, it inhibits and degrades BRAFV600E protein. Hence, we investigated the advantage of CRBN(BRAF)-24 as a degrader/inhibitor over a mere inhibitor. First, to determine whether CRBN(BRAF)-24 potently inhibits downstream signaling by degrading BRAFV600E, we compared ERK phosphorylation in BRAFV600E-positive cells treated with CRBN(BRAF)-24 and CRBN(BRAF)-29 in the presence or absence of a proteasome inhibitor, MG132. The active degrader CRBN(BRAF)-24 inhibited ERK phosphorylation more potently than the inactive degrader CRBN(BRAF)-29, but the better inhibitory effect of CRBN(BRAF)-24 was diminished by MG132 (Figure 6A). These results strongly suggest that CRBN(BRAF)-24 potently suppresses downstream ERK signaling by both inhibiting and degrading BRAFV600E.

As for the advantages of PROTACs, catalytic degradation of target proteins has been reported to result in sustained drug action.27, 38, 39 To examine the sustained suppression of ERK phosphorylation, cells were treated with CRBN(BRAF)-24, PLX8394, and CRBN(BRAF)-29 for 24 hours, and then further incubated in drug-free medium (Figure 6B,C). After 24 hours of drug treatment (washout 0 hour), BRAF protein was reduced only in the cells treated with CRBN(BRAF)-24. At this timepoint, CRBN(BRAF)-24 and PLX8394 at 100 nM almost completely inhibited the level of phosphorylated ERK in SK-MEL-28 cells, while CRBN(BRAF)-29 required a concentration that was 10 times higher to achieve equivalent inhibition (Figure 6B). The suppressed ERK phosphorylation was maintained for 24 hours in CRBN(BRAF)-24–treated cells even after drug removal but was gradually restored in cells treated with PLX8394 and CRBN(BRAF)-29, especially at low concentrations. Surprisingly, the level of BRAFV600E protein was further reduced in the CRBN(BRAF)-24–treated cells during 24 hours incubation in drug-free medium. In line with this, treatment of SK-MEL-28 cells with CRBN(BRAF)-24 for 24 hours effectively inhibited cell proliferation in drug-free medium for the subsequent 72 hours (Figure 6C). In a parallel experiment, PLX8394 and CRBN(BRAF)-29 had little effect. These results suggest that CRBN(BRAF)-24 shows potent and sustained suppression of downstream ERK signaling by degrading BRAFV600E, thereby inhibiting the proliferation of BRAFV600E-driven cancer cells more potently than kinase inhibitor.
4 DISCUSSION
The three currently FDA-approved RAF inhibitors (vemurafenib, dabrafenib, and encorafenib) provide significant benefits to patients with metastatic melanoma by potently inhibiting the MAPK pathway in tumors with BRAFV600 mutations.19, 40, 41 However, the emergence of acquired resistance seriously limits their clinical benefits.42, 43 Combination with MEK inhibitors neutralizes the paradoxical activation in BRAFWT cells caused by these RAF inhibitors and supports an improved therapeutic profile, but resistance and tumor recurrence remain inevitable.13 Therefore, there is the need for another approach to suppress oncogenic BRAFV600 mutants more potently and selectively. In this study, we successfully developed PLX8394-based PROTACs that potently induce the degradation of BRAFV600E protein in a CRL4CRBN E3 ligase–dependent manner. CRBN(BRAF)-24 degrades BRAFV600E at nanomolar concentrations, and the effect was sustained for more than 72 hours. Biochemical analysis revealed that BRAFV600E degradation by CRBN(BRAF)-24 induces a more potent and sustained inhibition of the downstream MAPK signaling than PLX8394, especially after drug removal. Thus, CRBN(BRAF)-24 is a potent BRAF degrader that outscores a BRAF inhibitor, PLX8394.
PLX8394 is a potent inhibitor of BRAFV600 mutant kinase (class 1) and can also disrupt both class 2 BRAF mutant homodimers and class 3 BRAF mutant heterodimers.36 Therefore, we further investigated the effects of CRBN(BRAF)-24 on the levels of BRAF mutant proteins, downstream kinase signaling and cell proliferation in cells endogenously expressing class 2 and class 3 BRAF mutants (Figures S3 and S4). CRBN(BRAF)-24 showed weak or minimal activity to reduce the class 2 and 3 BRAF mutants and did not inhibit kinase signaling in these cells except in NCI-H508 cells (Figure S3). Consistent with this, CRBN(BRAF)-24 did not effectively inhibit the proliferation of these cell lines (Figure S4). In NCI-H508 cells (heterozygous BRAFG596R, class 3), CRBN(BRAF)-24 showed significant BRAF-degradation activity (Figure S3) but did not inhibit cell proliferation (Figure S4), suggesting that the proliferation of this cell line may not be solely dependent on the RAF-ERK pathway.
The currently approved RAF inhibitors selectively inhibit monomeric BRAFV600 mutant proteins.13 However, these inhibitors (collectively known as group 1 RAF inhibitors) are known to enhance the formation of BRAF homodimers or heterodimers between BRAF and other RAF proteins,44 and binding of these inhibitors to one protomer of the RAF dimers results in a cooperative effect that activates the other protomer.13 This mechanism leads to reactivation of the MAPK pathway and promotes acquired resistance to BRAFV600 inhibition in cancer cells.36, 45, 46 Furthermore, the paradoxical activation of this pathway in normal cells underlies the development of proliferative skin lesions such as keratoacanthomas20, 21 and may therefore trigger the progression of secondary RAS-driven skin tumors.22, 47-50 Group 2 RAF inhibitors (e.g., BGB659, TAK632, LY3009120), which have recently been reported as effective inhibitors of RAF monomers and dimers, induce only weakly paradoxical activation, but their therapeutic window may be narrow because they inhibit ERK signaling and cell proliferation in normal cells as well as in cancer cells at concentrations effective for therapy.36 PLX8394 is one of the next-generation BRAF inhibitors, which are often referred to as “paradoxical breakers,” disrupting BRAF dimerization and tending to prevent both inhibition and paradoxical activation of the MAPK pathway in BRAFWT cells.13, 23, 36 In line with this, CRBN(BRAF)-24, which uses PLX8394 as a ligand, also induced neither inhibition nor paradoxical activation of ERK signaling in BRAFWT cells, even in the presence of oncogenic RAS mutations (Figure 4B) or upstream activation by EGF (Figure S5). Recently, other groups have reported CRBN-based PROTACs (P4B, compound 12, and 23) or a VHL-based PROTAC (SJF-0628), which incorporated vemurafenib or BI882370, that induce degradation of BRAF mutant proteins (mainly BRAFV600E) but not BRAFWT.51-53 However, these PROTACs have been shown to inhibit or paradoxically activate the MAPK pathway in BRAFWT cells, especially in cells with oncogenic RAS mutations or elevated upstream receptor signaling.51, 53 Therefore, CRBN(BRAF)-24 is the first selective BRAFV600E degrader that showed little effects on BRAFWT cells, a feature not observed in previous BRAF degraders.
The development of clinical resistance to RAF inhibitors in BRAFV600E-driven cancers is associated with the upregulated function of RAS, which acts by driving the drug-responsive monomeric form of BRAFV600E into the drug-unresponsive dimeric form with CRAF.54, 55 Posternak et al. reported that the transduction of oncogenic RAS mutant isoforms into A375 cells causes marked resistance to BRAFV600E degradation by a BI882370-based PROTAC, P4B.51 Similarly, we observed that A375 cells became resistant to CRBN(BRAF)-24 by the introduction of active RAS mutations (Figure S6B). However, BRAFV600E degradation by CRBN(BRAF)-24 in these cells was not affected (Figure S6A), suggesting that the BRAFV600E-degrading function of CRBN(BRAF)-24 is not suppressed by the activation of upstream RAS.
In summary, we have developed a new PROTAC against BRAFV600E protein by incorporating PLX8394 as a target ligand. CRBN(BRAF)-24 induced potent and selective degradation of BRAFV600E and exhibited antiproliferative activity against BRAFV600E-driven cancers without inducing paradoxical activation in BRAFWT cells. Although further optimization is required for its clinical development, CRBN(BRAF)-24 could be a novel lead as a BRAFV600E-targeted drug.
ACKNOWLEDGMENTS
We thank H. Nikki March, PhD, from Edanz (https://jp.edanz.com/ac), for editing a draft of this manuscript. We would also like to acknowledge Daiichi Sankyo RD NOVARE Co., Ltd. for the high-resolution mass spectrometry and NMR analysis and the preparation of the compounds.
DISCLOSURE
M. Suzuki, T. Uchida, M. Yoshida, and H. Ohki are employees of Daiichi Sankyo Co., Ltd. M. Naito is a member of the social cooperation program supported by Eisai Co., Ltd. and serves as a scientific advisor to UBiENCE Inc. M. Naito is an associate editor of Cancer Science. The other authors declare no conflict of interest. All authors had full access to all the data in the study and had final responsibility for the decision to submit this manuscript for publication.




