CXCR4 promotes the growth and metastasis of esophageal squamous cell carcinoma as a critical downstream mediator of HIF-1α
Xianxian Wu, Hongdian Zhang, Zhilin Sui, and Yongyin Gao contributed equally to this work.
Abstract
C–X–C motif chemokine receptor 4 (CXCR4) belongs to the CXC chemokine receptor family, which mediates the metastasis of tumor cells and promotes the malignant development of cancers. However, its biological role and regulatory mechanism in esophageal squamous cell carcinoma (ESCC) remain unclear. Here, we found that CXCR4 expression was associated with lymph node metastasis and a poor prognosis. In vitro and in vivo studies demonstrated that CXCR4 overexpression promoted ESCC cell proliferation, migration, invasion, and survival, whereas silencing CXCR4 induced the opposite effects. Mechanically, HIF-1α transcriptionally regulates CXCR4 expression by binding to a hypoxia response element in its promoter. HIF-1α-induced ESCC cell migration and invasion were reversed by CXCR4 knockdown or treatment with MSX-122, a CXCR4 antagonist. Collectively, these data revealed that the HIF-1α/CXCR4 axis plays key roles in ESCC growth and metastasis and indicated CXCR4 as a potential target for ESCC treatment.
Abbreviations
-
- CXCR4
-
- CXC chemokine receptor 4
-
- ESCC
-
- esophageal squamous cell carcinoma
-
- GV230
-
- HIF-1α scramble plasmid
-
- GV230-HIF-1α
-
- HIF-1α overexpression plasmid
-
- HIF-1α
-
- hypoxia inducible factor-1α
-
- HRE
-
- hypoxia response element
-
- KYSE30-fLuc
-
- KYSE30 cell line with the luciferase label
-
- oeCtrl
-
- CXCR4-overexpression control
-
- oeCXCR4
-
- CXCR4 overexpression
-
- oeHIF-1α
-
- HIF-1α overexpression
-
- oeNC
-
- HIF-1α-overexpression control
-
- shCtrl
-
- CXCR4-knockdown control
-
- shCXCR4
-
- CXCR4 knockdown
-
- shHIF-1α
-
- HIF-1α knockdown
-
- shNC
-
- HIF-1α-knockdown control
1 INTRODUCTION
Esophageal cancer (EC) is the sixth leading cause of cancer-related death worldwide, with an estimated 604 000 new cases and 544 000 related deaths globally in 2020.1 ESCC accounts for approximately 90% of all patients with EC in developing countries.2 Esophagectomy is a potentially curative treatment for patients with resectable ESCC. However, most patients present with locally advanced disease or distant metastases at the time of diagnosis, and these are the main reasons for poor outcomes.3, 4 Therefore, there is an urgent need to find potential biomarkers for the early diagnosis and treatment of ESCC.
CXC chemokine receptor 4 (CXCR4), a member of the G protein-coupled receptor (GPCR) family, consists of 352 amino acid residues and is expressed in hematopoietic stem/progenitor cells, pre-B cells, and endothelial cells.5, 6 CXCR4 was first identified as a coreceptor for human immunodeficiency virus type 1, and the gene is located on human chromosome 2q22.1.7 CXCR4 has been reported to be upregulated in at least 23 human cancer types, including pancreatic cancer, cervical cancer, and oral squamous cell carcinoma, in which it mediates tumor survival and metastasis.8-10 At present, many small-molecule CXCR4 inhibitors, such as AMD3100 for hematological malignancies,11 CTCE-9908 for osteosarcoma, and Olaptesed Pegol (NOX-A12) for myeloma and chronic lymphoblastic leukemia,12, 13 have been designed and approved by the US Food and Drug Administration for the treatment of patients. There is evidence suggesting that CXCR4 is overexpressed in ESCC and that CXCR4 overexpression enhances the migration and invasion of ESCC cells.14 However, the molecular regulatory mechanisms underlying CXCR4 overexpression in ESCC are not clear, and the role of CXCR4 in ESCC is not fully understood.
Hypoxia is an important biological parameter of solid tumors. Hypoxia inducible factor-1 (HIF-1) is the main regulator of hypoxia, and is composed of oxygen-dependent HIF-1α and constitutively expressed HIF-1β.15 Under normoxic conditions, HIF-1α is recognized by the von Hippel-Lindau protein for ubiquitin-mediated degradation, but under hypoxic conditions, HIF-1α is protected from degradation and dimerizes with HIF-1β to form the HIF-1 complex, transcriptionally activating several downstream effector genes.16 Tumor hypoxia and HIF-1α overexpression are involved in angiogenesis, survival, metastasis, metabolic reprogramming, and immune escape.17 HIF-1α has been reported to be highly expressed in ESCC and participates in the malignant tumor process.18 By screening the genomic DNA sequence of the human CXCR4 region, 7 hypoxia response elements (HREs) were found. On this basis, we hypothesize that CXCR4 may be an effective target of HIF-1α in ESCC.
In this study, we aimed to investigate (1) the role of CXCR4 in ESCC progression, (2) the mechanism by which HIF-1α regulates CXCR4, and (3) the correlation between HIF-1α and CXCR4 in ESCC specimens.
2 MATERIALS AND METHODS
2.1 Cell culture and reagents
Human ESCC cell lines EC109, KYSE140, KYSE30, KYSE150, KYSE450, KYSE510, and CaES17 were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The human normal esophageal epithelial cell line Het-1A was obtained from the American Tissue Culture Collection (ATCC). The human embryonic kidney cell line 293T and the luciferase-labeled KYSE30 cell line (KYSE30-fLuc) were kindly provided by colleagues in other departments. All cancer cells were cultured in RPMI 1640 (Gibco, Carlsbad, CA, USA) supplemented with 10% FBS (BI, Carlsbad, CA, USA). Het-1A and 293T cells were cultured in DMEM (Gibco, Carlsbad, CA, USA) containing 10% FBS. Cells were kept in an humidified incubator at 37°C and 5% CO2. Hypoxia (<1% O2) experiments were performed in a hypoxia incubator (Billups-Rothenberg Inc, USA). The CXCR4 antagonist MSX-122 was purchased from MedChemExpress (Shanghai, China).
ESCC cell lines were searched on Cellosaurus (https://web.expasy.org/cellosaurus/).19 Keyword C4024 (NCI Thesaurus Code for ESCC) was used for searching, and the data of ESCC cell lines used in our study are shown in Table S1. The Cancer Cell Line Encyclopedia (CCLE) is a database that contains multiomic data of over 1000 cancer cell lines, 28 ESCC cell lines included. Before analysis in this paper, cell lines from CCLE were checked to see if any problematic cell lines existed.
2.2 Clinical samples and immunohistochemistry (IHC)
Gene Expression Omnibus (GEO) data from the website (https://www.ncbi.nlm.nih.gov/geo/) were downloaded for further analysis.20 The expression of CXCR4 and HIF-1α was detected by IHC in 122 ESCC tissues and 28 adjacent noncancerous specimens. The clinical features of the patients are listed in Table 1. IHC analyses of the specimens were performed with anti-HIF-1α and anti-CXCR4 antibodies as described previously.21 Survival time was defined from the date of surgery to the date of death or the last follow-up. All operations for human subjects were approved by the Research Ethics Committee of our hospital, and informed consent forms were signed and provided by all patients.
| Clinicopathological features | Cases (n = 122) | Expression level | P-value | |
|---|---|---|---|---|
| CXCR4low (n = 52) | CXCR4high (n = 70) | |||
| Age (years) | ||||
| <68 | 58 | 22 | 36 | .318 |
| ≥68 | 64 | 30 | 34 | |
| Sex | ||||
| Male | 101 | 41 | 60 | .320 |
| Female | 21 | 11 | 10 | |
| Tumor size | ||||
| <3.5 cm | 48 | 20 | 28 | .863 |
| ≥3.5 cm | 74 | 32 | 42 | |
| Histological grade | ||||
| G1 | 16 | 9 | 7 | .147 |
| G2 | 99 | 42 | 57 | |
| G3 | 7 | 1 | 6 | |
| pT category | ||||
| T2 | 11 | 6 | 5 | .067 |
| T3 | 69 | 34 | 35 | |
| T4 | 42 | 12 | 30 | |
| Lymph node metastasis | ||||
| None | 72 | 36 | 36 | .048* |
| Yes | 50 | 16 | 34 | |
Note:
- *Statistically significant.
- Abbreviations: CXCR4, CXC chemokine receptor 4; ESCC, esophageal squamous cell carcinoma; G1, well differentiated; G2, moderately differentiated; G3, poorly differentiated/undifferentiated.
2.3 Quantitative real-time PCR (qRT-PCR) and RNA sequencing (RNA-seq)
Total RNA was isolated from tissues or cells with TRIzol reagent (Invitrogen, CA, USA), and cDNA was then synthesized using the PrimeScript™ RT Reagent kit (TaKaRa Bio, Shiga, Japan). Real-time PCR was performed with SYBR Premix Ex Taq II (TaKaRa Bio, Shiga, Japan). The fold-changes in expression were quantified using the 2−ΔΔCT method. Table S2 lists all primers used in this study. RNA-seq analysis was carried out by the Beijing Genomics Institute (Shenzhen, China).
2.4 Western blotting and enzyme-linked immunosorbent assay (ELISA)
Proteins from tissues or cells were extracted with protein lysate containing protease inhibitors. Western blotting was performed as previously described.21 The protein bands were detected by enhanced chemiluminescence. Table S3 lists all antibodies used in this study. The CXCL12 protein level in the cultured supernatants of ESCC cells was detected using an ELISA kit (MBBIOLOGY, Jiangsu, China) following the manufacturer's instructions.
2.5 Stable cell line construction
Lentiviral particles with CXCR4 overexpression (oeCXCR4) or CXCR4 knockdown (shCXCR4) were purchased from GenePharm (Shanghai, China). KYSE450 and KYSE510 cells were infected with oeCXCR4 or scrambled lentiviral particles (oeCtrl). Stably infected (KYSE450-oeCXCR4, KYSE450-oeCtrl, KYSE510-oeCXCR4, and KYSE510-oeCtrl) cell lines were obtained. KYSE30 and KYSE150 cells were infected with shCXCR4 or scrambled lentiviral particles (shCtrl). Stably infected (KYSE30-shCXCR4#1, KYSE30-shCXCR4#2, KYSE30-shCXCR4#3, KYSE30-shCtrl, KYSE150-shCXCR4#1, KYSE150-shCXCR4#2, KYSE150-shCXCR4#3, and KYSE150-shCtrl) cell lines were obtained.
Lentiviral particles with HIF-1α overexpression (oeHIF-1α) or HIF-1α knockdown (shHIF-1α) were purchased from GeneChem (Shanghai, China). KYSE450 and KYSE510 cells were infected with oeHIF-1α or scrambled lentiviral particles (oeNC). Stably infected (KYSE450-oeHIF-1α, KYSE450-oeNC, KYSE510-oeHIF-1α, and KYSE510-oeNC) cell lines were obtained. KYSE30 and KYSE150 cells were infected with shHIF-1α or scrambled lentiviral particles (shNC). Stably infected (KYSE30-shHIF-1α, KYSE30-shNC, KYSE150-shHIF-1α, and KYSE150-shNC) cell lines were obtained. Puromycin was used to screen all stably infected cell lines for more than 2 wk.
2.6 RNA interference and transfection
SiCXCR4 was synthesized by GenePharm (Shanghai, China). The sequences of siCXCR4 were as follows: forward 5′-CUGUCCUGCUAUUGCAUUATT-3′, reverse 5′-UAAUGCAAUAGCAGGACAGTT-3′. Subsequently, 2 × 105 cells/well were inoculated into a 6-well plate and transfected with 100 nM siRNA using Liposome 2000 (Invitrogen, CA, USA). After culture for 48-72 h, the cells were collected for subsequent experiments.
2.7 Cell proliferation assays
For the Cell Counting Kit-8 (CCK-8) experiment, 2 × 103 cells/well were inoculated into a 96-well plate and cultured for a specified time. The absorbance/well at 450 nm was measured using a microreader.
For the colony formation assay, 500 cells/well were seeded into a 6-well plate and cultured for 14 d. The colonies were fixed with 4% paraformaldehyde for 15 min and stained with crystal violet.
In the 5-ethynyl-2′-deoxyuridine (EdU) assay, the cells were inoculated into 24-well plates and cultured for 24 h. The proliferating cells were stained with EdU and counted by Leica DMI6000B.
2.8 Cell migration, invasion, and chemotactic assays
Transwell migration and Matrigel invasion assays were conducted with an 8-μm micropore chamber (Corning, NY, USA). Cells were suspended in serum-free RPMI 1640 and inoculated into the upper chamber, and RPMI 1640 containing 20% FBS was loaded into the lower chamber. After incubation for 24 h under hypoxic or normoxic conditions, migrated or invaded cells were fixed with 4% paraformaldehyde and stained with crystal violet. The chemotaxis assay was similar to the invasion assay, and RPMI 1640 containing CXCL12 (100 ng/mL) was loaded into the lower chamber as a chemoattractant.
2.9 In vivo studies
Male BALB/c nude mice (5-6 wk old) were purchased from HFK Bioscience Co. Ltd. (Beijing, China). Subsequently, 5 × 106 stably infected ESCC cells were injected subcutaneously into the right lateral axillary region. Tumor growth was monitored every 7 d, and the tumor volumes were calculated with the following formula: (length × width2)/2. The mice were sacrificed 28 d later, and tumor tissues were processed for further experimental.
In total, 5 × 105 stably infected KYSE30 cells labeled with luciferase (KYSE30-fLuc) were injected into the tail veins of mice to establish the lung metastasis model. The mice were divided randomly into 2 treatment groups and given either MSX-122 (10 mg/kg, daily, intraperitoneal injection) or PBS for 14 d consecutively. After 2 mo, the mice were injected intraperitoneally with fluorescein and imaged with a bioluminescence imaging system. The signal is displayed as photons/s/cm2/sr. Lung tissue was prepared for monitoring metastatic nodules. Animal experiments were approved by the ethics committee of our hospital for animal research (Ek2017027).
2.10 Dual-luciferase assay and chromatin immunoprecipitation (ChIP) assays
The HIF-1α overexpression plasmid (GV230-HIF-1α) was constructed by inserting PCR-amplified fragments into the GV230 vector (GeneChem, Shanghai, China). The wild-type, truncated, and mutant sequences of the CXCR4 promoter were amplified and cloned into the fLuc-GV238-basic vector (GeneChem, Shanghai, China). The resulting reporter plasmids were named GV238-1970 (−2002 to +92 bp, including HRE sites 1-7), GV238-1155 (−1200 to +92 bp, including HRE sites 1-5), GV238-296 (−500 to +92 bp, including HRE site 1) and GV238-mut (mutation of CXCR4 promoter HRE sites 1-7). Reporter plasmids and GV230-HIF-1α or GV230 were cotransfected into 293T cells under normoxia or hypoxia. A Renilla luciferase reporter was used as an internal control. The transcriptional activity of the cells was detected quantitatively using a dual-luciferase assay kit (Promega, WI, USA).
A ChIP commercial kit (Millipore, MA, USA) was used for the ChIP assay following the manufacturer's instructions. The primers used are listed in Table S2.
2.11 Statistical analysis
SPSS 16.0 and GraphPad Prism 7 software were used for data analysis. The quantitative data are presented as the mean ± standard deviation (SD) of at least 3 independent replicates. The differences between the 2 groups were analyzed by Student t test. The chi-square test was used to analyze the relationship between CXCR4 and clinicopathological variables. The survival curve was constructed by the Kaplan-Meier method and compared using the log-rank test. The correlation between the expression of HIF-1α and CXCR4 in ESCC tissue was analyzed by Spearman coefficient tests. Statistical significance was set at P < .05.
3 RESULTS
3.1 The expression of CXCR4 and its relationship with clinicopathological feathers and survival in ESCC patients
Based on publicly available data from the GEO, CXCR4 is highly expressed in ESCC tissues compared with normal tissues (GSE23400, GSE75241; Figure 1A). We also assessed CXCR4 expression by qRT-PCR and western blotting of fresh-frozen ESCC and adjacent nontumor tissues. CXCR4 was upregulated in ESCC tissues compared with normal tissues (Figure 1B,C). To further investigate CXCR4 expression in ESCC samples, immunohistochemical analysis was conducted to determine the expression of CXCR4 in 122 ESCC specimens and 28 esophageal normal mucosa tissues. A representative image is shown in Figure 1D. High expression of CXCR4 was found to be associated with lymph node metastasis (Table 1; P = .048). As shown in Figure 1E, the prognosis of patients with high expression of CXCR4 was significantly worse than that of patients with low expression of CXCR4 (P = .016). In addition, we determined the endogenous protein levels of CXCR4 in a group of ESCC cell lines and the immortalized esophageal normal cell line Het-1A. Western blotting analysis demonstrated that CXCR4 was highly expressed in KYSE30 cells with high metastatic ability, poorly expressed in KYSE450 and KYSE510 cells with low metastatic ability, and weakly expressed in Het-1A cells (Figure 1F). These results indicated that CXCR4 was upregulated in ESCC and may be used as a critical biomarker for patients with ESCC.
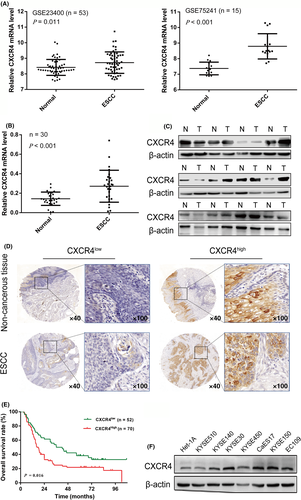
3.2 CXCR4 accelerates ESCC cell proliferation and growth
To validate the biological effect of CXCR4 among ESCC cell lines, KYSE30 and KYSE150 cells with relatively high expression of CXCR4 were selected for CXCR4 gene knockdown, and KYSE450 and KYSE510 cells with relatively low expression of CXCR4 were selected to construct stable CXCR4 overexpression cell lines. As shown in Figure 2A,B, compared with their respective controls, shCXCR4#2 significantly downregulated CXCR4 expression in KYSE30 and KYSE150 cells, and oeCXCR4 effectively upregulated CXCR4 expression in KYSE450 and KYSE510 cells. We then used shCXCR4#2 and oeCXCR4 for further research. KYSE30 and KYSE150 cells displayed attenuated proliferation following CXCR4 knockdown in short-term cell proliferation assays (CCK-8, colony formation, and EdU assays), whereas increased proliferation was observed in KYSE450 and KYSE510 cells with CXCR4 overexpression (Figures 2C,D and S1).
We also used a mouse xenograft model to verify whether CXCR4 affected the tumorigenesis of ESCC cells in vivo. The quantification of tumor volume showed that KYSE30 cells with stable CXCR4 knockdown generated smaller tumor masses than the control cells. Conversely, tumor growth was increased in mice injected with KYSE450 cells overexpressing CXCR4 compared with mice injected with the corresponding control cells (Figure 2E).
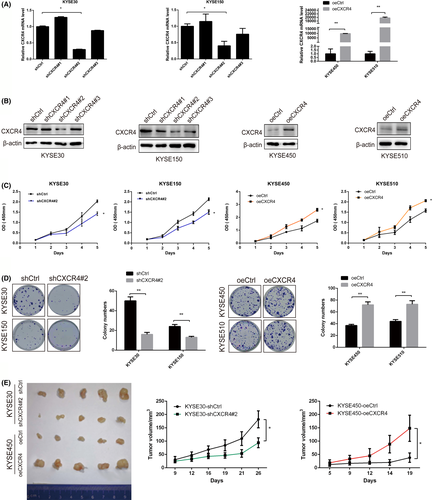
3.3 CXCR4 promotes ESCC cell migration and invasion
Transwell experiments were conducted to determine the potential role of CXCR4 in regulating the migration and invasion of ESCC cells. CXCR4 knockdown significantly reduced the migration and invasion ability of KYSE30 and KYSE150 cells (Figure 3A), whereas CXCR4 overexpression induced the opposite effects in KYSE450 and KYSE510 cells (Figure 3B).
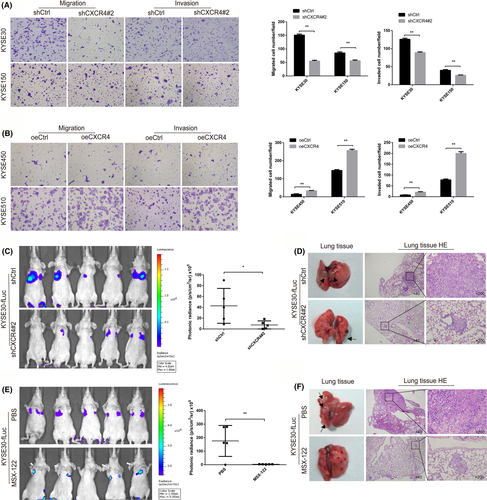
A mouse xenograft model was used to verify whether CXCR4 affected the metastasis of ESCC cells in vivo. Stable KYSE30-fLuc transfectants (KYSE30-fLuc-shCtrl and KYSE30-fLuc-shCXCR4) were implanted into nude mice by tail vein injection. As expected, CXCR4 knockdown reduced the lung metastasis of KYSE30 cells, which was monitored by an in vivo luciferase imaging system (Figure 3C,D). To verify the anticancer properties of MSX-122 (a CXCR4 antagonist) in vivo, a KYSE30-fLuc mouse xenograft model was constructed. The mice were divided randomly into 2 treatment groups; they were given either MSX-122 (10 mg/kg, daily, intraperitoneal injection) or PBS for 14 d consecutively and then fed for 2 mo. The results demonstrated that MSX-122 inhibited the lung metastasis of KYSE30 cells in vivo (Figure 3E,F).
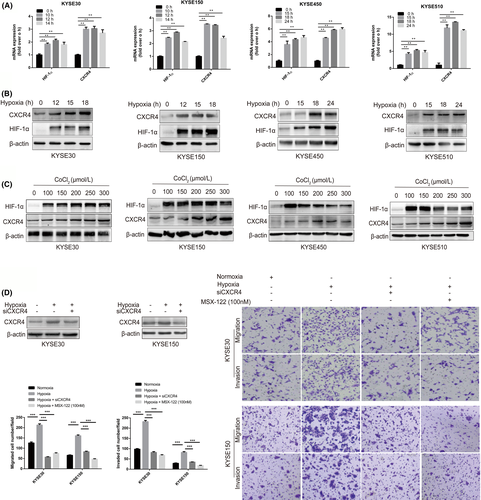
3.4 CXCR4 expression is upregulated under hypoxic conditions in ESCC cells
To estimate whether CXCR4 expression could be induced by hypoxia, we exposed KYSE30, KYSE150, KYSE450, and KYSE510 cells to hypoxic conditions (1% O2). The CXCR4 mRNA and CXCR4 protein levels increased markedly under hypoxic conditions (Figure 4A,B). The expression of CXCR4 was also detected in ESCC cells treated with CoCl2, a known HIF-1α stabilizer. CoCl2 upregulated CXCR4 protein expression in the 4 ESCC cell lines (Figure 4C). Furthermore, an in vitro transwell assay demonstrated that the increased migration and invasion abilities of KYSE30 and KYSE150 cells induced by hypoxia could be weakened by CXCR4 interference or treatment with MSX-122 (100 nM) (Figure 4D).
3.5 Close relationship between HIF-1α and CXCR4 expression in ESCC cells
Given that HIF-1α is the main regulator of oxygen homeostasis, we detected the transcriptome of KYSE30-shNC and KYSE30-shHIF-1α cells using RNA-seq analysis. Further functional enrichment analysis revealed a group of target genes significantly related to cancer-related functions (Figure 5A). CXCR4 has been shown to be a gene that responds to hypoxia and participates in the regulation of cell migration and invasion (Figure 5B). Subsequently, we verified the effect of altered HIF-1α levels on the expression of CXCR4 in ESCC cells. As shown in Figure 5C,D, HIF-1α knockdown in KYSE30 and KYSE150 cells markedly reduced the mRNA and protein levels of CXCR4 under normoxia or hypoxia, whereas HIF-1α overexpression upregulated CXCR4 expression in KYSE450 and KYSE510 cells (Figure 5E). We further investigated the CXCL12 protein levels in the culture supernatant of ESCC cells. ELISA detection showed that the expression of CXCL12 protein was not induced by HIF-1α (Figure 5F), which may support the theory that cancer cells expressing CXCR4 are directionally transferred to target organs with high CXCL12 secretion.
Given the critical roles of HIF-1α and CXCR4 in ESCC progression, IHC analysis of HIF-1α expression was conducted using the same ESCC specimens used for CXCR4 staining. High expression of HIF-1α was found to be associated with lymph node metastasis (Table S4; P = .013) and poor prognosis of ESCC (Figure S2; P =.028). Representative images are shown in Figure 5G. HIF-1α protein levels in ESCC tissues were positively correlated with CXCR4 expression (Figure 5H; R = .54, P < .001). Moreover, the GEO data showed a positive correlation between HIF-1α and CXCR4 expression (Figure 5I).
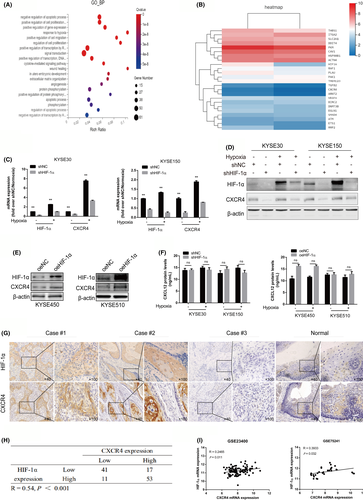
To verify the important role of CXCR4 in HIF-1α-mediated ESCC cell migration and invasion, we interfered with CXCR4 expression in KYSE450 and KYSE510 cells overexpressing HIF-1α. As shown in Figure 6A, transwell assays showed that downregulation of CXCR4 expression weakened the promoting effect of HIF-1α overexpression on the migration and invasion of KYSE450 and KYSE510 cells. As expected, MSX-122 treatment (100 nM) also attenuated the migration and invasion ability of the 2 ESCC cell lines induced by HIF-1α.
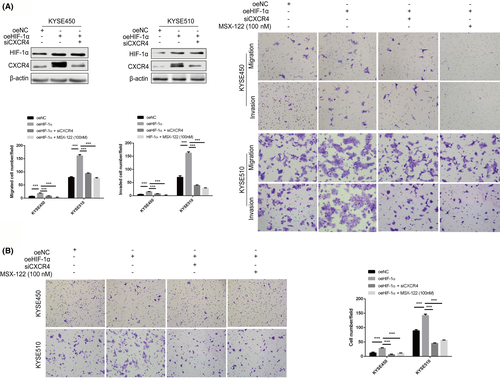
CXCL12/CXCR4-mediated chemotaxis promotes the directional metastasis of CXCR4-expressing cancer cells toward organs with high CXCL12 protein secretion. Therefore, we explored further whether CXCR4 mediated the chemotaxis of ESCC cells induced by HIF-1α toward CXCL12. The experimental results suggested that ectopic HIF-1α expression promoted the chemotaxis migration of KYSE450 and KYSE510 cells toward CXCL12 (100 ng/mL), but this effect could be blocked by CXCR4 interference or MSX-122 treatment (100 nM) (Figure 6B).
3.6 HIF-1α regulates CXCR4 expression in ESCC cells
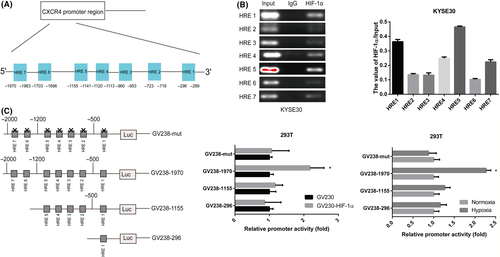
To explore the regulatory mechanism of CXCR4 overexpression in ESCC, we analyzed the CXCR4 promoter sequence and found 7 putative HREs (Figure 7A). ChIP assays revealed that HIF-1α directly bound to HRE sites 1, 4, 5 and 7 in the CXCR4 promoter in KYSE30 cells under hypoxic conditions (Figure 7B). In addition, we constructed a CXCR4 luciferase promoter reporter plasmid GV238-1970 (containing HRE sites 1-7), truncated reporter plasmids GV238-1155 (containing HRE sites 1-5) and GV238-296 (containing HRE site 1), and a mutant reporter plasmid GV238-mut. We cotransfected a reporter plasmid with or without HIF-1α overexpression into 293T cells. Dual-luciferase analysis demonstrated that HIF-1α significantly enhanced GV238-1970 transcriptional activity in 293T cells, but HIF-1α upregulation did not activate the transcriptional activity of these truncated or mutated reporter genes. We further verified that the transcriptional activity of GV238-1970 was enhanced under hypoxic conditions. As expected, hypoxia did not activate the transcriptional activity of these truncated or mutated reporter genes (Figure 7C). Therefore, we concluded that HIF-1α regulated CXCR4 transcription by binding to the HRE site in the promoter region of the CXCR4 gene.
4 DISCUSSION
Esophageal cancer is one of the most common and lethal malignancies in China and worldwide. Despite considerable advances in surgical techniques and comprehensive treatment, the prognosis of patients remains poor because of early recurrence and metastasis.22 Therefore, a better understanding of the molecular mechanisms of ESCC tumorigenesis and the identification of biomarkers are of great importance for improving the effectiveness of early diagnosis and treatment for ESCC.23
Increasing numbers of studies have shown that CXCR4 is highly expressed in a variety of cancers, including colorectal cancer, breast cancer, and gastric cancer.24-26 However, the biological role and potential regulatory mechanism of CXCR4 in ESCC remain unclear. In this study, we found that the expression of CXCR4 was upregulated in ESCC cell lines and tissue specimens. High expression of CXCR4 was found to be related to lymph node metastasis and a poor prognosis. CXCR4 is a transmembrane receptor; however, our IHC results revealed that CXCR4 was mainly expressed in the membrane and cytoplasm of ESCC cells. This phenomenon may be because CXCR4 is internalized during activation and transferred from the membrane to the cytoplasm, therefore resulting in specific downstream signals. Biological studies have shown that CXCR4 gene knockdown decreased ESCC proliferation, migration, and invasion in vitro, and growth and metastasis in vivo, whereas overexpression of CXCR4 induced the opposite effects. These results strongly supported the role for CXCR4 as a protumor gene in ESCC.
Cancer immunotherapy has shown strong anticancer activity in a variety of cancers. Clinical trials have suggested that PD-1 inhibitors are an attractive target for the treatment of ESCC.4, 27 Although anti-PD-1 therapy provides a novel direction for ESCC, there are still some limitations, such as low immunogenicity.27 Researchers have identified CXCR4 as a promising target for cancer immunotherapy. Li et al28 suggested that blocking CXCR4 reduced tumor fibrosis to improve anti-PD-L1 immunotherapy. Inhibition of CXCR4 improved immunotherapy in breast cancer.29, 30 In this research, CXCR4 was proven to be tumorigenic in ESCC. Blocking CXCR4 had a potential inhibitory effect on the metastasis of ESCC. However, there was no evidence regarding the efficacy of CXCR4 combined with PD-1 inhibitors in the treatment of ESCC.
Accumulating evidence has proven that CXCR4 has tumorigenicity and preliminarily determines its carcinogenic mechanism. The interaction of CXCR4 and CXCL12 activates G protein-dependent signaling pathways with broad effects on promoting tumor survival and metastasis.31 Studies have also reported that homodimerization of CXCR4 induced the G protein-independent JAK/STAT pathway, which is involved in chemotactic responses.32 In addition, Pan and colleagues found that the transcription factor FOXC1 increased the expression of CXCR4, boosting breast cancer metastasis.33 Hong et al34 proposed that vascular endothelial growth factor (VEGF) upregulates the expression of CXCR4 to promote the invasion of glioma cells. Another study reported that epidermal growth factor (EGF) enhanced the expression of CXCR4, thereby promoting the metastasis of malignant cells in lung cancer.35 However, the potential mechanism of CXCR4 in regulating ESCC biology remains unclear.
Increasing evidence has suggested that hypoxia is an important driving force for tumor progression. HIF-1α, the main regulatory factor of hypoxia, is thought to induce the expression of multiple target genes, which is beneficial to the tolerance of tumor cells to hypoxia.36, 37 In this study, we found that the expression of CXCR4 was significantly increased under hypoxic conditions (1% O2). In addition, the expression of CXCR4 was induced by HIF-1α, and HIF-1α-enhanced cancer cell motility was weakened by the downregulation of CXCR4 expression or treatment with MSX-122. HIF-1α activated the transcriptional expression of CXCR4 by binding to its promoter HRE region in ESCC cells. These results suggest that HIF-1α is involved in the upregulation of CXCR4 expression in ESCC cells. However, several limitations must be mentioned. First, the tissue sample size involved in this study was relatively small. Second, the detailed molecular mechanisms and signaling pathways by which HIF-1α/CXCR4 influence the malignant progression of ESCC were not investigated. Finally, there is no discussion about the clinical transformation of this pathway in ESCC. Therefore, future research is still needed to address these issues.
In conclusion, we demonstrated the important role of CXCR4 in promoting ESCC growth and metastasis. HIF-1α enhances the transcriptional activity of CXCR4 by binding to its promoter HRE sites. The current research data suggest that HIF-1α/CXCR4 may serve as a potential therapeutic target for ESCC patients.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81772619, 82002551), the Science and Technology Project of Tianjin Municipal Health Commission (KJ20119), the Bethune Charitable Foundation-Excelsior Surgical Fund (HZB-20190528-11, HZB-20190528-18), and the Clinical Trial Project of Tianjin Medical University (2017kylc006).
DISCLOSURE
The authors declare that they have no conflict of interest.




