Scientific history, biogeography, and biological traits predict presence of cryptic or overlooked species
ABSTRACT
Genetic data show that many nominal species are composed of more than one biological species, and thus contain cryptic species in the broad sense (including overlooked species). When ignored, cryptic species generate confusion which, beyond biodiversity or vulnerability underestimation, blurs our understanding of ecological and evolutionary processes and may impact the soundness of decisions in conservation or medicine. However, very few hypotheses have been tested about factors that predispose a taxon to contain cryptic or overlooked species. To fill this gap, we surveyed the literature on free-living marine metazoans and built two data sets, one of 187,603 nominal species and another of 83 classes or phyla, to test several hypotheses, correcting for sequence data availability, taxon size and phylogenetic relatedness. We found a strong effect of scientific history: the probability of a taxon containing cryptic species was highest for the earliest described species and varied among time periods potentially consistently with an influence of prevailing scientific theories. The probability of cryptic species being present was also increased for species with large distribution ranges. They were more frequent in the north polar and south polar zones, contradicting previous predictions of more cryptic species in the tropics, and supporting the hypothesis that many cryptic species diverged recently. The number of cryptic species varied among classes, with an excess in hydrozoans and polychaetes, and a deficit in actinopterygians, for example, but precise class ranking was relatively sensitive to the statistical model used. For all models, biological traits, rather than phylum, appeared responsible for the variation among classes: there were fewer cryptic species than expected in classes with hard skeletons (perhaps because they provide good characters for taxonomy) and image-forming vision (in which selection against heterospecific mating may enhance morphological divergence), and more in classes with internal fertilisation. We estimate that among marine free-living metazoans, several thousand additional cryptic species complexes could be identified as more sequence data become available. The factors identified as important for marine animal cryptic species are likely important for other biomes and taxa and should aid many areas in biology that rely on accurate species identification.
I. INTRODUCTION
Cryptic species (CS) are reproductively isolated groups belonging to the same nominal species, often because of a lack of morphological differences between them (Struck et al., 2018). There are two types of CS: CS sensu stricto (CS ss), where there are truly no morphological differences between CS, and pseudo-CS, representing overlooked species (Collins & Cruickshank, 2014), where reproductively isolated groups share a nominal species, even though there are morphological differences between the groups (Chenuil et al., 2019). Although it is often important to distinguish between cases of CS ss and pseudo-CS (e.g. understanding of evolutionary phenomena leading to one or the other, such as natural selection on morphological features), in other cases it is not practical (e.g. assessing the extent to which biodiversity estimates are wrong, or for biological monitoring which may require non-experts to identify species rapidly in the field). In addition, establishing an absence of morphological differences depends on the characters studied, the instruments available and the methods used, so it can be very difficult to conclude whether a taxon contains CS ss. We therefore use CS to refer to both cases herein, unless otherwise stated.
CS are being identified at an increasing rate due to the use of genetic markers (Knowlton, 1993, 2000; Struck et al., 2018). Several articles have argued that the identification of CS could dramatically change global biodiversity estimates (Adams et al., 2014; Li & Wiens, 2022), improve connectivity estimates (Hubert et al., 2012; Pante et al., 2015; Cerca, Purschke & Struck, 2018; Sheets, Warner & Palumbi, 2018) and shed light on evolutionary processes (Struck et al., 2018; Chenuil et al., 2019), for example by correcting diversification rate estimates (Faurby, Eiserhardt & Svenning, 2016; Utami et al., 2022). Unidentified CS often directly impact agriculture, medicine (Pérez Ponce de León & Nadler, 2010) and assessments of ecosystem vulnerability (Chenuil et al., 2019). Knowlton (1993) conducted a large-scale review of marine CS. Many CS were morphologically or ecologically differentiated, so CS often reflected lack of precision in morphospecies definition (i.e. they were overlooked species or pseudo-CS). She argued that the abundance of marine CS related to the difficulty of accessing this habitat, and predicted that organisms relying on chemical cues for mating were more likely to contain CS than those relying on visual cues. The focus on CS was later extended to non-marine organisms (Bickford et al., 2007). Although an early quantitative analysis emphasised the generality of CS presence across taxa and regions (Pfenninger & Schwenk, 2007), this data set revealed a difference of two orders of magnitude in the frequency of CS among metazoan taxa (Trontelj & Fišer, 2009). This was confirmed by Pérez-Ponce de Leon & Poulin (2016) who highlighted that well-studied groups also contained CS. Poulin & Pérez-Ponce de León (2017) refuted the hypothesis that the number of CS per species complex was higher for parasites than free-living organisms, but they found a higher number for freshwater than marine, and to a lesser extent terrestrial, taxa.
Struck et al. (2018) discussed the difficulty of correctly inferring the presence of CS and found that many of the 606 papers surveyed did not contain sufficient data to identify CS (e.g. to establish reproductive isolation). Fiser, Robinson & Malard (2018) reviewed 120 amphipod papers and found that CS were generally not recently diverged, suggesting that natural selection (niche conservatism or parallelism) may be responsible. Chenuil et al. (2019) proposed a framework to guide the study of CS and detailed how to detect and classify CS and identify their multiple causes, which may relate to human taxonomic activity (species definition) and eco-evolutionary processes (natural selection, or neutral polymorphism allowed by high effective population sizes).
Despite these reviews, and with the exception of Poulin & Pérez-Ponce de León (2017), no hypotheses have been statistically tested to explain which features are associated with CS abundance. This question is timely since ecologists, evolutionists, biogeographers and biodiversity managers all need to know how many species have been overlooked. Overlooking CS may lead to inaccurate estimates of community diversity, or of distribution range or abundance within a species (Chenuil et al., 2019; Muir et al., 2022). Identifying high-level patterns of CS presence will allow researchers and managers to predict where one might find cases of CS and how biased biodiversity data are (e.g. Shin & Allmon, 2023).
CS could be caused by three types of factors (Chenuil et al., 2019): (i) the quality of the taxonomic work for species delineation and description; (ii) recent divergence among sister species; and (iii) eco-evolutionary processes like natural selection or genetic drift for example. These factors led us to choose candidate variables that are likely to influence the probability of the presence of CS in a nominal species, or their abundance within a taxonomic class (Table 1). First, the quality of taxonomic work depends on the technologies available when the species was described or on the presence of suitable morphological characters in the taxon. This may be especially true for genitalia, which are known to be related to reproduction and thus reproductive differences. Conversely organisms with external fertilisation often rely on chemical cues for conspecific mating, potentially providing fewer characters for taxonomy and leading to more CS (Knowlton, 1993). In the absence of clear morphological differences, and when CS are in allopatry and controlled crosses are not feasible, molecular tools are the only solution to establish reproductive isolation. The generalisation of simple molecular tools and universal markers, beginning in the 1980s, may have contributed to a reduction in the number of CS among nominal species described during the last few decades. Additionally, the nature of the prevailing theory of living organisms may influence taxonomists due to differences in geographical distribution range, niche width and morphology expected within a species. Second, recent speciation may explain morphological similarity and thus the presence of CS (particularly CS ss). Third, eco-evolutionary processes like stabilising natural selection, but also genetic drift (neutral models) could lead to CS ss or pseudo-CS. This category is potentially very broad; we chose to investigate the specific hypothesis that natural selection to avoid hybridization may accelerate morphological differentiation among sister species that rely on visual cues to mate, so fewer CS may be expected in classes with image-forming vision (Mayr, 1963; Bickford et al., 2007).
| Nominal species data set | Class-level data set | |||||||
|---|---|---|---|---|---|---|---|---|
| Latitude zone | Year | Genus size | Distribution range | Hard skeleton | Vision | Fertilisation | External genitalia | |
| Quality of taxonomic work | ||||||||
| Theory of evolution | x | x | ||||||
| Morphological diagnosis constraints: | ||||||||
| - technological | x | |||||||
| - taxon morphology | x | x | ||||||
| Difficult to assess reproductive isolation: | ||||||||
| - absence of molecular tools | Refuted | |||||||
| Recent speciation | x | x | ||||||
| Eco-evolutionary process | ||||||||
| Selection against hybridization | x | x | x | |||||
| Level of significance | NA | *** | n.s. | *** | **/* | */n.s.# | */* | n.s./n.s. |
- The effect of the different variables was tested on the probability of presence of CS in a nominal species (left four variables) or on CS abundance in a taxonomic class (right four biological variables). Statistical significance determined by the current study (see Section III): ***, P < 0.001; **, P < 0.01; *, P < 0.05; #, P < 0.10; n.s., not significant. Significance values before/after the ‘/’ symbol correspond to models without and with phylogenetic correction, respectively. The hypothesis that an absence of molecular tools when species were described led to more CS cases was refuted, since early-described species have more sequence data available (see Table 3, Fig. S2).
To test these associations and to search for global predictors of CS using variables that are available for many metazoan species, we performed a systematic survey of free-living marine metazoan CS from the literature and identified 977 taxa containing CS. Crossing these data with other publicly available information described in Section II.2, we built two data sets: one containing information on geography, taxonomic variables and sequencing effort in nominal species, and the other containing biological traits compiled at the taxonomic-class level. We tested our predictions using generalised linear models (GLMs) or generalised estimating equation models (GEEs) correcting for phylogenetic relatedness and considering whether nucleotide sequence data were available or not.
To test our predictions related to the timing and quality of taxonomic work, we obtained the year of species description for marine metazoan species, as well as distribution range data when possible. At the class level, we gathered information on the presence of a hard skeleton and external genitalia because they can provide morphological characters for taxonomy, as well as fertilisation mode (internal or external). Relating to recent speciation, and since quaternary glaciations triggered recent speciation, we compared latitudinal regions that were differentially affected by glaciation (CLIMAP, 1981). We also investigated whether large genera were associated with CS, given that genera with many species may reflect high recent diversification rates. Lastly, we used class-level data to test the hypothesis that organisms with image-forming vision have fewer CS. We specify that, although stabilising selection on morphology, or convergence, is often hypothesised to explain particular CS cases, this cannot be tested at the meta-analysis level with the present data.
II. METHODS
(1) Cryptic species data set and literature survey
Our literature search was conducted using Web of Science, surveying articles published through the end of 2020. We used key words pertaining to marine metazoan species, journals that publish marine research, CS, and both genetic and morphological or other phenotypic data. The full set of key words can be found in the online Supporting Information, Appendix S1. Additionally, we performed a search in Web of Science to identify all studies published through the end of 2020 that cited Knowlton (1993) or Knowlton (2000). These records were combined and duplicates were removed; we then scanned the abstracts to remove papers that did not identify CS in marine metazoans. Papers whose abstracts were unclear were read in full to determine if the study should be included or not (Fig. 1). We chose to focus our study on free-living marine metazoan species (both sessile and mobile animals), eliminating studies on algae and parasites.
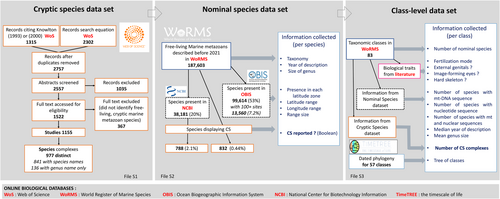
After removal of duplicates, filtering and abstract screening, the initial list of 2757 distinct scientific articles was screened for relevance. The remaining 1522 studies were read in full to determine if they provided information on free-living marine metazoan CS. 1155 suitable articles were identified, corresponding to 977 distinct taxa which were ultimately retained for statistical analyses (Fig. 1). We generally did not re-evaluate the authors' original conclusions about species status (i.e. whether CS were present or not) except for some cases where we split multiple CS complexes within a genus after careful reading of the article and analysing phylogenetic relationships (see Section III). As a rule, we tried to be conservative to avoid overestimating the number of CS (Leaché et al., 2019; Chan et al., 2022).
Many papers identified more than one nominal species with CS. When we could clearly identify two (or more) different nominal species containing CS within a single genus based on a single paper, we entered these CS separately into our data set. When this was not possible, CS were entered into our database as part of a genus. Species that were reported as CS in more than one paper were only retained once in the database. The full CS data set is available as File S1 in the permanent repository (see Section VII or IX).
(2) Data sets derived from public databases
We constructed two data sets (nominal species data set and class-level data set) using publicly available databases and our CS data set, then used them to test hypotheses about CS. We used basic R tools unless specified otherwise. Species names were standardised and updated among all data sets based on the World Register of Marine Species database (WoRMS) which contains information on both accepted and unaccepted names (Costello et al., 2013). The main variables and statistical analyses are detailed below and summarised in Appendix S2.
(a) Nominal species data set and analyses
This data set was built to test whether geographical and taxonomic variables or sequencing effort influenced the probability of CS being present. We first created a data set of all marine metazoan nominal species by filtering the WoRMS database (January 2021 version). We retained species in Animalia, with taxonomic status ‘accepted’, and flagged as ‘extant’ and ‘marine’. We eliminated the following entirely parasitic taxa: Acanthocephala (phylum), Dicyemida (phylum), Neodermata (superclass in the phylum Platyhelminthes), Myxozoa (class in the phylum Cnidaria) and Tantulocarida (class in the phylum Arthropoda). We computed genus size (i.e. number of congeners) for each species based on this filtered list.
We then queried NCBI GenBank (https://www.ncbi.nlm.nih.gov/) using a custom-designed Perl script (available in the permanent data repository, see Section VII or IX) to find, for each species in the filtered WoRMS list, the number of nucleotide sequences and the number of mitochondrial sequences present in GenBank (NCBI) published before the end of 2020. To evaluate the availability of genetic data for each species, we transformed the sequence counts to Boolean variables ‘ncbi’ (= presence of nuclear nucleotide sequence data in NCBI), ‘mito’ (= presence of mitochondrial nucleotide sequence data in NCBI), and ‘mitonuc’ (= presence of both nuclear and mitochondrial sequence data in NCBI) which were ‘true’ when there was at least one DNA sequence available (or for ‘mitonuc’, one nuclear and one mitochondrial sequence). This reduction was appropriate because identical sequences are seldom submitted more than once to GenBank even if they are sequenced from several different individuals. Therefore, species with CS may have more unique sequences in NCBI than those without CS which could lead to circularity. We also built a categorical variable for the type of nucleotide sequences available for a species (none, nuclear only, mitochondrial only, both types).
We computed genus size: the number of nominal species in our final data set for each genus. We extracted the year of species description from the WoRMS database field ‘scientificNameAuthorship’ which had fewer missing data than the field ‘yearPublishedIn’.
To obtain geographical information, we searched the Ocean Biodiversity Information System (OBIS) (www.obis.org) using the R package robis (Provoost & Bosch, 2017). We divided the globe into five latitudinal zones: North Polar (90° N – 66.5° N), North Temperate (23.5° N–66.5° N), Tropical (23.5° N–23.5° S), South Temperate (23.5° S–66.5° S), and South Polar (66.5° S–90° S). We queried OBIS to see if each nominal species occurred within each zone (presence/absence only). To control for differences in study effort among species, we rarefied the distribution range data: we retained only species with occurrences from at least 100 distinct sites in OBIS, with random sampling of 100 sites when more were available, to compute distribution ranges. We extracted the ranges of latitude and longitude (as the maximum minus the minimum) and used the convex hull estimator for distribution range using the package GeoRange (Boyle, 2017). The convex hull estimator does not exclude land between sampling sites and therefore may overestimate marine range sizes. Although the rarefaction procedure will preferentially remove species with smaller distribution ranges, it was necessary for conservative distribution range comparisons: indeed, species with CS generally had many more sites reported in OBIS before rarefaction (median number of sites 157 versus 6 for species without CS, the average numbers of sites being 8.5-fold higher with CS). This higher number of sites (which may be partly due to higher study effort, not only to broader distribution) may artificially inflate the mean distribution range of species with CS, which was about sevenfold larger than species without CS before rarefaction. Since this ratio is much lower after rarefaction (see Section III.1), it can be considered a conservative estimate since species with lower distribution ranges were preferentially removed.
We completed the data set with the presence of CS (present in our CS data set: yes/no). The full nominal species data set contained 187,603 nominal species described before 2021, with presence in each latitude zone for 53% of these, and distribution ranges for only 7.23% (13,560 species) (Fig. 1). Nine species with CS were not included in the nominal species data set because we did not find an accepted name in WoRMS (April 2020 version) or the accepted name in WoRMS did not correspond to any entry (as ‘scientificName’) with an ‘accepted’ status. Therefore, there are 832 nominal species with CS in the nominal species data set (File S2 in the permanent data repository, see Section VII or IX) but 841 in our survey (File S1 in the permanent data repository, see Section VII or IX) (Fig. 1). We did not reintegrate these species by manually correcting our WoRMS database subset for fear of biasing the data, since their particular taxonomic status may reflect that they were under revision and may no longer be considered as CS.
We used logistic regression to model the probability that CS would be detected within a nominal species (Boolean variable: ‘has CS’) using the following explanatory variables: a sequencing effort variable (type of sequence available in NCBI: ncbi, mito, mitonuc; see Table 2), a variable for time of species description (year or historical period), distribution range, and a taxonomic variable (phylum or ‘phylum_class’ which is a concatenation of phylum and class corresponding to the 83 taxa in the class-level data set; see Fig. 1 and Appendix S2). We chose time periods based on landmark publications by Darwin (1859), Mayr (1942), and Kimura (1979, 1983), which divided the study into four time periods (1759–1859; 1860–1942; 1943–1980; 1981–2020). We also performed our analyses for three periods, either removing Mayr's breakpoint (1942), or removing Kimura's breakpoint (1980) (see Appendix S2). We searched for the best model of logistic regression using the ‘glm’ function in R or the ‘bestglm’ function of the bestglm R package (McLeod, Xu & McLeod, 2020), which compared models including some or all of these explanatory variables. To determine the best model, we used the Bayesian Information Criterion (BIC), which is generally more conservative than the Akaike's Information Criterion (AIC) regarding the inclusion of additional explanatory variables. Model selection was also carried out without the geographic variables to include all nominal species, since a large percentage of species did not have geographic information in our data set (see above).
| No sequence data | Nuclear only | Mitochondrial only | Nuclear and mitochondrial | Total with sequence data in NCBI | |
|---|---|---|---|---|---|
| Total (with and without reported CS) | 149,450 | 3145 | 11,971 | 23,037 | 37,153 |
| CS not reported | 149,406 | 3,131 | 11,836 | 22,398 | 36,365 |
| CS reported | 44 | 14 | 135 | 639 | 788 |
| % with CS/total | 0.03 | 0.45 | 1.13 | 2.77 | 2.12 |
Since year of species description had a significant influence on the probability of the presence of CS, we used a method to find, without a priori expectations, the times when CS excess (‘calibrated’ by sequencing effort) changed, by grouping years into decades. For this, we computed the residuals of a logarithmic regression of the number of CS per decade (identity function, Poisson distribution, zero intercept) against the number of species with NCBI data. We detected changes in means for these residuals using the Bayesian change point method in the bcp package (Wang & Emerson, 2015). We also used this package to infer whether particular decades corresponded to a change in average species distribution ranges since changes in evolutionary biology theory may predispose authors to describing species with larger or smaller distribution ranges.
Using information from each study, we identified cases for which reproductive isolation among CS was established unambiguously, i.e. sympatric CS that were studied using codominant markers or several independent markers (Chenuil et al., 2019), or cases with morphological, ecological or reproductive differentiation among CS (Folder D in the permanent data repository, see Section VII or IX), although further analyses of these factors were not carried out. This allowed us to check that our results were not biased by authors' decisions to report CS based on a priori hypotheses (e.g. on distribution ranges): we checked that significant variables identified in the best GLM model to explain CS presence remained significant when including as CS only unambiguous cases of reproductive isolation.
(b) Class-level data set and analyses
This additional data set was built with the aim of testing whether CS are more abundant in some groups due to their biological traits. Biological trait information was available at the class level, but was not reliable at the nominal species level, due to numerous possible exceptions and missing data. We collapsed the WoRMS species list to the class level, building a data set (class-level data set) of 83 marine metazoan taxa (Fig. 1). For phyla without classes, we considered the entire phylum. For each class (or phylum), we reported the number of nominal species containing CS based on our literature survey. For this variable, which is our dependent variable in subsequent analyses, we also included the 136 taxa containing CS which were not identified to the species level but for which we had their genus name. For each class (or phylum), we calculated the number of accepted extant marine species in WoRMS, mean genus size, and median year of description. Additionally, based on the NCBI data (see Section II.2.a), we calculated the number of species containing at least one nucleotide sequence in NCBI, the number of species containing at least one mitochondrial nucleotide sequence and the number of species with both nuclear and mitochondrial sequences. For this analysis by class, we used the April 2020 version of WoRMS (instead of January 2021, as for other analyses) because the newer version included the class Hexanauplia, which has since been invalidated. Hexanauplia united Copepoda (mobile species with image vision) with Thecostraca (sessile species without vision), and both taxa have since been rehabilitated to the class level; we included them as separate classes in our analysis.
Using Brusca, Moore & Shuster (2016), we added the following biological variables to the class-level data set: fertilisation mode (internal/external/both) or the related Boolean variable (internal fertilisation vs. external/both), presence of external genitalia (yes/no/both), presence of image-forming eyes, and the presence of a hard skeleton. These variables are generally fixed at the class level, but the inclusion of ‘both’ as an option allowed us to account for classes with a significant proportion of both internal and external fertilisers (e.g. within the class Gastropoda, phylum Mollusca), or both internal/external genitalia. We chose these variables based on a priori ideas of how scientists define different species and how species may be morphologically differentiated in the field.
To understand which variables influenced the number of nominal species with CS in a class, we used a two-step analysis. The first step consisted of fitting a GLM with a Poisson distribution and identity link function, of the number of CS cases in a class, with explanatory variables representing class size and effort in terms of DNA sequencing. The second step used the residuals of the first-step model, which represent the excess in CS cases after controlling for taxon size and study effort, and we investigated the effect of taxonomic or biological variables on these residuals. For this second step, we performed two distinct analyses. The first was a linear model analysis retaining all 83 classes or phyla; the second analysis corrected for phylogenetic relatedness among classes or phyla and was therefore restricted to the subset of 57 classes or phyla for which a timed phylogenetic tree was available. This second analysis had the advantage of accounting for phylogenetic relatedness but the disadvantage of a smaller sample size reducing statistical power.
For the first step, we hypothesised that the number of CS detected in a class depended on the availability and type of genetic data and taxon size. We thus computed distinct models to obtain the residuals (res0, res1, res1m, res1b, or res2, in File S5 in the permanent data repository, see Section VII or IX), which represent the excess of CS when class (or phylum) size and sequencing effort are accounted for. The first of these models (computing res0) was analogous to a model used in previous studies (Pfenninger & Schwenk, 2007; Trontelj & Fišer, 2009). The other models differed in the variables used to account for sequence data availability. Since results were very similar among the five residual types, we only present results for the model with residuals res1b which gave the best AIC. For this model, to obtain res1b, we fitted a Poisson regression with a null intercept and an identity link function, to explain the number of CS in a class or phylum, by the number of species for which both nuclear and mitochondrial sequences were available, and the number of species in the class. The residuals (res1b) were then compared among classes/phyla to identify taxa in which CS may be particularly prevalent and to assess a possible influence of biological traits. We used analyses of variance to compare linear models (based on BIC or AIC) with a single biological variable and with taxonomic variables (mean genus size, median year of species description for the class, and phylum) to determine the extent to which variation in the residuals (excess of CS cases) could be explained (class-level data set, N = 83).
To perform statistical analyses correcting for phylogenetic relatedness among the 83 classes, we obtained a phylogenetic tree with dated divergence times from timetree.org (Kumar et al., 2017), containing 57 metazoan classes. Tips were pruned or renamed using the ape package (Paradis & Schliep, 2019) to harmonise taxonomic nomenclature between timetree.org and WoRMS without modifying topology or divergence times. We then filtered the data set so it contained only the 57 classes for which phylogenetic information was available using ‘compar.gee’ in the ape package (R). This function performs the comparative analysis using GEE (Paradis & Claude, 2002). This analysis incorporates a correlation matrix that specifies the dependence among observations, which is obtained from the phylogenetic tree. We used the residuals of the previous Poisson regression (res1b) to analyse the excess or deficit of CS with compar.gee (specifying a Gaussian distribution). We then tested each biological variable one by one in a linear model.
III. RESULTS
Our literature search yielded 1155 studies of free-living, marine metazoans corresponding to 977 CS complexes (see Fig. 1). The survey data set is available as File S1 in the permanent data repository (see Section VII or IX). 841 of these were identified as nominal species, although nine were then removed because they did not correspond to a valid ‘scientificName’ entry in WoRMS, resulting in a total of 832 nominal species. The remaining 136 CS complexes were reported only as genera, as we were not able to assign a species name to a CS complex. This occurred for instance when a study (often a molecular phylogenetics study) identified several divergent genetic groups contradicting the monophyly of at least two nominal species identified by their morphology.
Different molecular markers were used to delineate species (File S1, counted using File S4, both in folder C, data repository, see Section VII or IX). Out of the 977 distinct species complexes detected, 40.1% were characterised by both nuclear and mitochondrial data, 46.5% only by mitochondrial data, and 12.2% only by nuclear data. The remaining 1.1% were described by other methods such as RAPD. Allozymes were available in 12.2%, microsatellites in 4.2%, and SNPs or omics data in only 2.0% of cases.
96 studies detected several distinct complexes of CS, up to a maximum of 33, often in barcoding studies or phylogenetic studies [e.g. Carr et al. (2011) detected 33 CS in Polychaeta; Moura et al. (2018) detected at least 29 CS in Hydrozoa]. These papers represented 368 CS complexes out of 977. Some CS complexes appeared in up to seven validated studies included in our survey [e.g. the actinopterygian fish Mugil cephalus (seven studies), the anthozoan Actinia equina and the nematode Litoditis marina (six studies each)], although we ultimately removed these duplicated studies prior to statistical analysis. 609 papers identified only a single CS complex. Since barcoding or phylogenetic studies had lower sample sizes than population genetic or phylogeographic studies, we checked our analyses using only these 609 cases; our main conclusions were upheld. For each CS case, File S1 in the online data repository (see Section VII or IX) contains the number of distinct CS cases found in each paper and the number of articles describing CS in each taxon.
(1) Factors influencing the probability of CS within a nominal species
Our WoRMS subset contained 187,603 accepted free-living metazoan nominal species from 30 phyla and 83 classes, of which 832 (0.44%) displayed CS. When sequence data were present in NCBI for a nominal species, the probability that CS were reported increased (Table 2). From the 19.8% of species that had nucleotide sequences in NCBI (37,153), 2.12% displayed CS and the proportion reached 2.77% for species with both mitochondrial and nuclear sequences. However, 5.29% (44 nominal species) of the 832 taxa displaying CS had no sequence data in NCBI (Table 2), generally because these CS reports were based on allozymes or microsatellites or because authors did not publish the nucleotide sequences. Geographical data were found for 99,614 species for which we could record presence in the five latitude zones, and distribution ranges were computed following rarefaction for the 13,560 species which occurred in at least 100 different sites in OBIS.
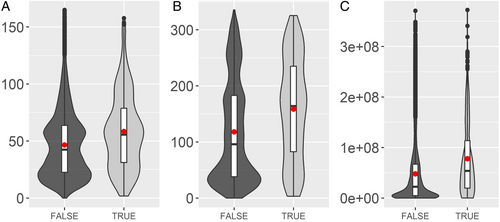
Species with reported CS had larger distribution ranges on average: 1.25-fold larger for latitudinal range, 1.34-fold for longitudinal range and 1.62-fold for the convex hull range estimate (Fig. 2, all three Wilcoxon rank-sum tests comparing species with CS and species without CS were highly significant, P < 10−14).
Time of species description strongly impacted the probability of reported CS. Both the number of nominal species described per year and the proportion of CS detected (either based on all species or based only on species with sequence data) varied through time. The earliest-described species tend to have higher proportions of CS, but this proportion did not decrease regularly (Fig. 3A). Bayesian change point analysis on CS excess per decade (computed as residuals of a regression of number of species per decade against the number of species with sequence data) suggests that a decrease in probability of CS occurs for species described after 1841–1850 and that this decrease becomes amplified in later decades, thus after publication of Darwin (1859) (Fig. S1). In nearly all major phyla, species with CS tended to be described earlier (by 62 years, on average) (Fig. 3B).
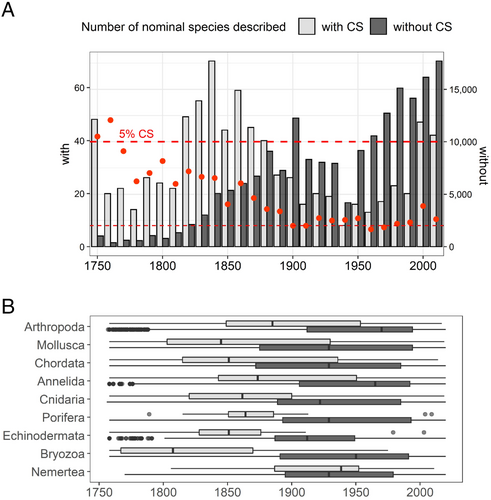
Other parameters varied with time of species description (Fig. S2). In particular, the proportion of nominal species for which sequence data are available decreased regularly for species described between 1800 and 1920 and then stabilised after 1950, with possibly a slight increase for species described after 2000 (Fig. S2A). In addition, species described in the 20th century tend to have smaller distribution ranges, with these ranges decreasing regularly with year of description since ca. 1950 (Fig. S2C, Table S1). For average distribution ranges sizes, Bayesian change point analysis detected a maximum and high probability of change (P = 0.8) after the decade 1921–1930, i.e. preceding the decade when Mayr (1942) was published (Fig. S3). Table 3 summarises the data for the four historical periods delimited by the major evolutionary biology publications Darwin (1859), Mayr (1942) and Kimura (1979, 1983). We investigated the differences between consecutive periods using GLM models explaining the probability of CS presence by both the presence of sequences in NCBI and historical period. We found a decrease in probability of CS for species described between 1756–1859 and 1860–1942 (‘Darwinian transition’; P < 0.001), between 1860–1942 and 1943–1980 (‘Mayr transition’; P < 0.01) and, when considering only three longer periods (i.e. removing the Mayr breakpoint), there was a non-significant increase between 1860–1980 and 1981–2020 (P = 0.0713) (R script File S5, folder D, public repository, section VII or IX). When distribution range was included in the model, the Darwinian transition remained highly significant, but the Mayr transition was no longer significant.
| Period | 1756–1859 | 1860–1942 | 1943–1980 | 1981–2020 |
|---|---|---|---|---|
| Number of nominal species (NS) | 20,978 | 67,485 | 35,915 | 63,225 |
| Number of NS with cryptic species (CS) (%) | 388 (1.85%) | 258 (0.38%) | 54 (0.15%) | 132 (0.21%) |
| Number of NS in NCBI (%) | 11,107 (52.95%) | 14,105 (20.90%) | 4,557 (12.69%) | 8,384 (13.26%) |
| Number of NS with at least 100 sites in OBIS | 6259 | 5670 | 1049 | 582 |
| Number of NS in NCBI & with 100 sites in OBIS | 5388 | 4074 | 652 | 338 |
| Number of NS with CS in NCBI (%) | 382 (3.44%) | 240 (1.70%) | 49 (1.07%) | 117 (1.39%) |
| Mean NS distribution range (million km2) | 51.85 | 51.67 | 33.47 | 18.81 |
- NCBI, National Center for Biotechnology Information; OBIS, Ocean Biogeographic Information System.
According to the Bayesian Information Criterion (BIC), the best model to explain the probability that a nominal species contained CS included three variables, all highly significant: type of nucleotide sequence available (none, nuclear, mitochondrial or both) (P < 10−15), distribution range (convex hull) (P < 10−13), and year of species description (P < 10−6). The taxonomic variables ‘phylum_class’ or ‘phylum’, when forced into the best model, were highly significant (P < 10−15) but the BIC increased by 28% or 6% respectively, so these models were far from optimal. For comparison, the maximum BIC difference between all models involving one variable for sequencing effort (among three possible variables), one for distribution range (among three possibilities) and one for description time (among four possible variables) was only 1.4% (Table S2). ‘Genus size’ was not in the best models; when added, it had a positive although non-significant coefficient. Because geographic distribution ranges were not available for 92.8% of the species following rarefaction, we also tested models without geographic variables and found the same overall results (P < 10−15 for each of the two variables retained). Using the less-stringent AIC criterion instead of the BIC resulted in more variables in the best model. In addition to year of species description, distribution range and DNA sequence availability, period of species description (with three periods: 1756–1859, 1860–1942, and 1943–2020), and ‘phylum’ were retained. All these variables were highly significant (P < 10−10) except ‘year of species description’ which was very significant (P < 0.01) and is obviously related to period of description.
We confirmed these results in analyses including only the 446 cases (54% of reported CS cases) for which reproductive isolation was unambiguously established. Year of species description (P < 10−6) and type of sequence data available (P < 10−15) remained highly significant predictors of CS presence, and distribution range became very significant (P = 0.0016). For species described during the 20th century, the correlation of distribution ranges with time of description (Fig. S2C) suggested a risk of collinearity, making it difficult to separate the effects of scientific history (date of description) and distribution range. However, variance inflation factors were lower than 1.02 (thus refuting collinearity), so we are confident that both year and distribution range independently influence the probability of the presence of CS (i.e. for a given distribution range, nominal species containing CS tended to be described earlier and for a given decade, they tended to have larger ranges, Fig. S4).
To investigate whether CS were more likely to occur in some latitudinal zones, we compared the proportions of CS, range sizes, and median year of description for each zone. The proportion of CS cases among species with sequence data was highest for species in the north polar zone (5.13%), followed by the south polar zone (3.57%), and lowest in the tropics (2.64%) (Table 4). Species from the north polar zone were described earlier (median description year 1872) and had the most abundant geographic data (45% of species had 100 sites or more in OBIS) yet distribution ranges were average; species from the south polar zone had the largest mean distribution range (92 million km2, based on 655 species with rarefied data) (Table 4).
| North polar | North temperate | Tropical | South temperate | South polar | |
|---|---|---|---|---|---|
| Number of nominal species (NS) | 4145 | 48,166 | 56,847 | 32,269 | 3384 |
| Number of NS containing cryptic species (CS) (%) | 108 (2.61%) | 609 (1.26%) | 575 (1.01%) | 418 (1.30%) | 41 (1.21%) |
| Number of NS in NCBI (%) | 2045 (49.3%) | 19,778 (41.1%) | 21,187 (37.3%) | 11,917 (36.9%) | 1063 (31.4%) |
| Number of NS with at least 100 sites in OBIS | 1884 (45%) | 10412 (22%) | 9846 (17%) | 7410 (23%) | 655 (19%) |
| Mean distribution range (million km2) | 65.782 | 58.421 | 60.545 | 72.081 | 92.441 |
| Median number of OBIS sites before rarefaction | 89 | 15 | 11 | 18 | 22 |
| Median description year | 1872 | 1905 | 1913 | 1909 | 1914 |
| Number of NS with CS in NCBI (%) | 105 (5.13%) | 588 (2.97%) | 560 (2.64%) | 405 (3.40%) | 38 (3.57%) |
- NCBI, National Center for Biotechnology Information; OBIS, Ocean Biogeographic Information System.
- We did not use rarefied data to allocate species to different latitudinal zones, thus we report median numbers of sites before rarefaction to compare coverage among zones.
The proportion of CS was much higher among the 162 pan-polar species (i.e. species reported from both the south and north polar zones), with 11.1% of the species with data in NCBI reporting CS. This was approximately five times the overall proportion found among metazoans (2.12%, or 2.42% when restricted to those with data in OBIS). Antitropical species (i.e. those present in Northern and Southern Hemisphere but not in the tropics; 1443 species) also have an increased proportion of CS (3.57% of their species in NCBI report CS) relative to that in all metazoans (see above), although a lesser extent than panpolar species.
(2) Effect of biological factors on the number of CS cases in a class
Our class-level data set was composed of 83 free-living taxa: 77 classes and 6 phyla which did not contain accepted taxonomic classes in WoRMS. Taxon size, number of species with sequences in NCBI, and number of species with mitochondrial DNA sequences, or with both nuclear and mitochondrial DNA sequences in NCBI, all had highly significant (P < 10−15) effects on the number of CS reported. Among the five models that used taxon size and sequencing effort (see Section II.2.b) to predict the number of CS in a class, the best model (with the lowest AIC = 573.17, for the other four models AIC = 579.82–581.39) was a multiple regression using the number of nominal species and the number of species with both nuclear and mitochondrial sequences available in NCBI (P < 10−10 and 10−15 for these explanatory variables, respectively). The ranking of classes based on the residuals from these models (and thus the ‘excess CS’ for a given research effort) depended slightly on the model (File S3 in the permanent data repository, see Section VII or IX) but the results of statistical analyses investigating which biological or taxonomic factors influenced the number of cases of CS were similar for all types of model used (see below).
We then analysed how the residuals of the best model were explained by biological traits, mean genus size, median year of species description, and phylum. When tested alone, phylum was not significant (although it was significant at P < 0.05 with the other types of residuals as dependent variables). In order to identify the variable that explained the most variation in the residuals, we sought the best model with a single explanatory variable (using bestglm). Phylum explained the least variation [BIC = 193.7, an increase of 59% compared with the best model and 48% compared with the second-worst model]. The best model included the presence of a hard skeleton (BIC = 125.5, P < 0.01, fewer CS in classes with hard skeletons), followed by image-forming vision (BIC = 128.15, P < 0.05, fewer CS in classes with image vision), then the Boolean variable ‘internal fertilisation’ (BIC = 128.4, P < 0.05, fewer CS in classes with internal fertilisation). Models including the following variables resulted in a higher BIC value than the null model (BIC = 128.5): fertilisation mode (internal/external/both; P < 0.05 despite its high BIC (BIC = 129.05), more CS in classes with both types of fertilisation), genus size (BIC = 132.7), median year of species description (BIC = 132.78), presence of external genitals (BIC = 132.96) and phylum (all non-significant). Assessment using the AIC criterion also identified the model containing phylum as the worst model.
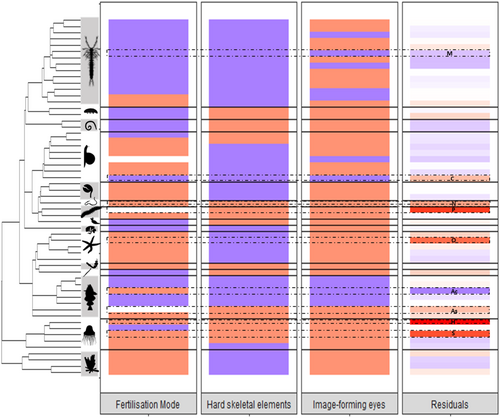
In the analysis correcting for phylogenetic relatedness with the data set of 57 classes for which a dated phylogeny was available (Fig. 4), two biological factors remained significant as single variables: fertilisation mode (P < 0.05) and presence of a hard skeleton (P < 0.05). When the analysis was carried out with CS ss (i.e. for CS with no diagnostic morphological differences present), all results were similar except that there was weak evidence for an association between image-forming vision and CS presence (P = 0.0811). We also investigated whether the influence of biological traits, or mean genus size or median year of description depended on the type of study, confirming that analysis of CS from studies only finding a single CS complex did not affect the results. Similarly, using only CS with confirmed reproductive isolation did not change the conclusions of our statistical analyses. When contrasting classes with internal fertilisation against all other classes pooled together, the effect of fertilisation mode was not significant in the phylogenetically corrected analysis. For both data sets, models predicted an excess of CS for classes with both types of fertilisation, relative to classes with only internal fertilisation or, to a lesser extent, external fertilisation, and fewer CS for classes with hard skeletons or with image-forming vision.
IV. DISCUSSION
(1) Some animal classes have more chances than others of having CS
We identified 977 cases of CS in marine metazoans and combined this database with publicly available data on molecular sequencing effort, geographical information, and biological traits, to allow us to identify significant predictors of the presence of CS. A key aspect of our study was the use of NCBI data to correct for sequencing effort. Together with distribution range and year of description, type of nucleotide sequence available had an extremely significant effect on the presence of CS. Taxa with CS excesses or deficits differ from those identified in previous studies (Trontelj & Fišer, 2009; Perez-Ponce de Leon & Poulin, 2016). We found a highly significant effect of taxonomic group in the nominal species data set analyses, although class or phylum were not included in the best models, suggesting that we identified the variables responsible for a large part of these taxonomic differences. Our finding of substantial excess CS in Polychaeta confirms previous results for Annelida (Trontelj & Fišer, 2009; Nygren, 2014) and the observed deficit of CS in Actinopterygians confirms trends from a previous study not restricted to marine species (Trontelj & Fišer, 2009). However, we refute that Porifera also displays an excess of CS or that Cephalopoda displays a deficit (Trontelj & Fišer, 2009). Our second key innovation was the inclusion of geographical, scientific history, and biological variables in our models, which proved significant CS predictors. Although the precise ranking of classes based on the residuals (i.e. excess of CS after accounting for sequencing effort and taxon size) vary slightly with the model used (residuals in File_S3, folder A, public repository, see Section VII or IX), our main conclusions on the historical, geographical and biological factors influencing them are stable across our analyses.
(2) Large distribution ranges are associated with CS
Nominal species with CS had, on average, 1.62-fold larger convex hull distribution ranges than other species. Considering that rarefaction eliminated species with fewer than 100 reported sites, and that average and median site numbers (before rarefaction) were higher for species with CS (see Section II.2.a), this ratio may be an underestimate. We recognise that this may represent a publication artifact. First, scientists may be more willing to interpret the presence of divergent groups of haplotypes as CS when they know the species has a wide distribution. However, we also found that distribution range affects CS probability when considering only CS with unambiguous reproductive isolation. Second, species with larger distributions may have been the subject of phylogeographic or population genetic studies more often, and thus benefitted more from NCBI data (and thus CS detection) than others. However, distribution range and presence in NCBI were both considered in our models and their variance inflation factors were negligible, implying that the effect of distribution range on CS presence is unlikely to be a consequence of the availability of sequence data.
The larger distribution ranges in nominal species with CS (Fig. 2) apparently supports the suggestion of Knowlton (1993) that molecular studies usually reveal that ‘cosmopolitan’ species are species complexes. However, CS complexes often include sympatric biological species each of which have large distribution ranges (Nygren, 2014; Egea et al., 2016; Brasier et al., 2017; Castelin et al., 2017; Fiser et al., 2018; Moura et al., 2018). We therefore cannot rule out that species with large distribution ranges may be more likely to form CS, which could result from multiple potential mechanisms. (i) Large distribution ranges may reflect ecological success and such species may have higher rates of recent diversification (Chenuil et al., 2018), generating novel species that have diverged too recently to be morphologically differentiated. This is supported by a positive (although non-significant) influence of genus size on the probability of CS. (ii) Alternatively, species with large geographic distribution ranges will have larger effective population sizes (a measure that is important in evolutionary processes and is directly linked to genetic diversity). They may thus evolve diagnostic morphological differences more slowly under a neutral model of morphological evolution, due to their longer coalescence times (Egea et al., 2016; Chenuil et al., 2019) leading to CS. (iii) Processes such as allopatric speciation or adaptation to different climate regimes may be more important in species with large distribution ranges. (iv) This finding is also compatible with a selectively neutral hypothesis predicting that old species had more time to experience geographic speciation, compared to young species. Testing this will require species-level data on taxon age or time since divergence, which was beyond the scope of the present study. Future analyses of more detailed characteristics of each CS case (e.g. morphology, allopatry or sympatry, ecology, number of CS in the complex) could shed light on some of these potential mechanisms.
(3) CS are in excess at the poles, not in the tropics
The proportions of CS per nominal species, even after accounting for study effort, are highest at the poles and lowest in the tropics (Table 4), contradicting the hypothesis that the tropics, as understudied zones of high biodiversity, will shelter more CS (Bickford et al., 2007) and that extreme biomes like polar zones will have fewer CS (Adams et al., 2014). A higher proportion of CS in the polar zones could be explained by shorter divergence times among sister species, since these areas were highly impacted by Pleistocene glacial cycles that triggered recent speciation. Divergence time among sister species is indeed longer, and recent speciation rates lower, in the tropics (Schluter, 2016). This latitudinal pattern of CS prevalence suggests that the latitudinal diversity gradient, with species richness highest at low latitudes, may be less steep than is currently thought, and contrasts with a recent analysis based on terrestrial birds, which also included CS (Freeman & Pennell, 2021). As with explanations of distribution range, the evolutionary age of species and habitats will be valuable information to explain CS patterns across latitudes.
Simple explanations invoking only the concentration of taxonomists or molecular biology facilities in northern temperate zones are unlikely to explain the observed CS latitudinal pattern because the northern temperate zone has an intermediate CS proportion comparable to the southern temperate zone. However, the fact that on average, north polar species were described earlier, and south polar species have larger distribution ranges than species of other latitudes, suggests that additional analyses will be necessary to interpret the observed latitudinal patterns.
(4) Scientific history matters
Year of species description strongly affected the probability of a taxon having CS. To interpret this pattern, we first envisage a null hypothesis to explain the decrease in CS probability with shorter times since description (Fig. 3). Whatever the prevailing scientific views and technical tools available, the first species described might have been the most abundant ones because it is natural to start grouping the most abundant objects when sorting things. Abundant species tend to have broader distributions (Holt et al., 1997; Gaston, Blackburn & Lawton, 1997) which increases the probability of CS presence. Another non-exclusive explanation is that early taxonomists had less-accurate tools available to describe species, leading to lumping of CS into a single nominal taxon, as well as a restricted view of Earth's biodiversity that might have assumed fewer species existed overall.
However, other factors may be at play because our analyses identified decreases in CS probability, corrected by sequencing effort, during the decades following the landmark publication of Darwin (1859) on natural selection (Fig. S1). The same method for average distribution ranges detected a decrease in distribution range both before and after the decades when Mayr (1942) was published (Fig. S3). In our comparison of four periods a priori defined by major evolutionary theories, we a found significant influence of two transitions on the excess of CS corrected by sequencing effort: a decrease in CS after Darwin (1859), a decrease after Mayr (1942), and a non-significant trend for an increase after Kimura (1979, 1983).
There are multiple possible explanations for this pattern. Early taxonomists did not consider species as evolving entities, regarding them as fixed until the early 19th century following Lamarck's theory of transformism (de Lamarck, 1809). Darwin's theory of evolution, relying on speciation via adaptation to different habitats, perhaps led taxonomists to recognise that species were restricted to particular environments, and thereby reduced the extent of species lumping. The smaller decrease in proportion of described species containing CS after Mayr (1942) may be explained by his argument for geographical isolation leading to divergence among populations, and the temporal coincidence of the observed decrease in distribution range decrease supports this view (Table 3). Furthermore, when including distribution ranges in the model, the Mayr transition no longer affected the probability of CS presence, suggesting that this decrease in CS was explained by distribution range decrease alone. Although we initially assumed that Mayr (1942)'s influence may have caused a decrease in distribution ranges of species described thereafter, the strongest decrease in decadal average distribution range in fact preceded Mayr (1942). This suggests instead that an increased number of geographically restricted species described may have influenced Mayr's understanding of life history and speciation. Although the availability of molecular markers in recent times should contribute to a greater correspondence between nominal species and biological species, the probability of having CS appears to have increased slightly for species described after 1980, particularly since 2000 (Fig. S2A). However, contrary to expectations, the proportion of species with sequence data was the lowest for species described post-Kimura (1979, 1983) and is highest for the earliest-described species (i.e. 1756–1859, see Table 3), perhaps supporting the view that the species described earliest were globally the most conspicuous, and thus most likely to be studied using later DNA sequencing methods. Alternatively, it is possible that changes in views close to publication of Kimura's neutralist theory of evolution (Kimura, 1979, 1983) influenced taxonomists towards accepting more morphological variation within species, assuming that it may not necessarily reflect differential adaptations. Hennig's (1966) development of cladistics theory with phylogenies based only on shared derived characters probably also influenced taxonomists, alongside arguments against over-splitting and poor-quality taxonomic works with limited sampling (Sangster & Luksenburg, 2015). The fewest CS were described during the period 1943–1980 after Mayr (1942) defined the biological species concept (Table 3). Our analyses identified a strong influence of the timing of species descriptions but assessing the role of these or other theories will require dedicated specific historical studies of the scientific process.
Additionally, historical patterns of species discovery and delineation may differ among taxonomic groups. For instance, polychaete taxonomy was greatly affected by three major 20th-century taxonomists who tended to lump earlier taxa together (Hutchings & Kupriyanova, 2018). This may explain the observed overabundance of CS in polychaetes, and there may be similarly idiosyncratic explanations for patterns in other taxa (e.g. Shin & Allmon, 2023).
(5) Some biological traits are associated with variations in CS abundance between classes
At the class level, the presence of hard skeletal components was associated with fewer CS. This agreed with our prediction that hard parts may be more reliable taxonomic characters for species description and can be measured and quantified in living and non-living specimens. Interpreting the effect of fertilisation mode requires more caution. Although the coefficient associated with the presence of both internal and external fertilisation was the highest in analyses correcting for phylogenetical relationships, only two of the three classes that show both fertilisation types have more CS than expected based on taxon size and sequence data, Polychaeta (phylum Annelida) and Ascidiacea (phylum Chordata). The third class with both fertilization types (Mollusca: Gastropoda) has a slight deficit in CS (see Fig. 4). Thus it is risky to generalise a result supported by only two classes. External fertilisation was associated with more CS, and internal fertilisers with fewer CS (particularly Arthropoda: Malacostraca) relative to expectations (albeit not significantly in the phylogenetically corrected analysis).
One explanation for the relative lack of CS in internal fertilisers is that fertilisation in these species often involves external genitalia. As with hard skeletal elements, genitalia are often used by taxonomists to distinguish species [e.g. isopods (Ribardiere et al., 2017); littorinid snails (Hohenlohe & Boulding, 2001)], and may be key in conspecific mate identification. For several CS in our data set, researchers used morphological differences in genitalia to distinguish CS or putative CS. However, the variable ‘presence/absence of genitalia’ was not significant, so this factor alone cannot account for the effect of fertilisation mode. Variations in modes of reproduction and larval type between CS of a species complex are known for various classes like ophiuroids (Weber, Stohr & Chenuil, 2014; Weber, Stöhr & Chenuil, 2019), asteroids (Knowlton, 1993; Hart, 1996; Hart, Byrne & Johnson, 2003; Naughton & O'Hara, 2009), gastropods (Ellingson & Krug, 2006), scleractinians (Schmidt-Roach et al., 2013) and zoanthids (Soong, Shiau & Chen, 1999), although these studies rarely document whether fertilisation is internal or external. Hydrozoans, the class with the most CS relative to expectations (Fig. 4), often have complex life cycles with both benthic and pelagic phases, as well as sexual and asexual reproduction. This may add a level of complexity to species delimitation in this group relative to those with simpler life histories, but detailed analysis is beyond the scope of the present study.
We also found potential support for the hypothesis that selection against heterospecific mating has triggered morphological differentiation among species with good vision, limiting the incidence of CS. More insight could be gained by genotyping or sequencing populations of cavernicolous or abyssal species that have lost the ability to form images to investigate whether CS are more frequent than in their relatives with image-forming eyes.
V. CONCLUSIONS
- (1)
We found support for the three categories of CS causes: (i) strong support for a role of taxonomic history (early species description) and morphological features (absence of a hard skeleton); (ii) indirect support for a role of recent speciation (latitudinal gradient and genus size trend); and (iii) weak support for a role of selection to avoid heterospecific mating (presence of image-forming eyes decreases CS probability).
- (2)
We found that it is likely that historical definitions of species entities are potentially biased by how researchers defined and detected species at the relevant times, arguing for the development and improvement of population genetics or population genomics in studies of biodiversity.
- (3)
Based on the present number of free-living metazoan species in WoRMS (187,603) and the 2.77% of species with nuclear and mitochondrial sequence data containing CS, we estimate that 4364 CS cases (5196, or 2.77% of the total number of species in WoRMS, minus the 832 cases that we identify here) are still undiscovered in free-living marine metazoans. Since only a few specimens were used for a high proportion of the species included in the NCBI database and only slowly evolving markers have been sequenced in numerous cases, this is likely to be an underestimate of the true number of CS complexes. In addition, we found indications that reports of CS may have begun to accelerate in recent years (Fig. 3). Similarly, when study effort increases, many known species complexes often reveal additional CS, as was the case in polychaetes (Nygren et al., 2018; Teixeira, 2022; Teixeira et al., 2022a,b, 2023).
- (4)
Currently undescribed CS may therefore be concentrated among species which were described before the first half of the nineteenth century and have wide distributions and no hard skeleton, particularly in polar zones.
- (5)
Although this study focuses on marine metazoans, we expect the historical, geographic, and biological factors identified as important could be applied to other taxa and biomes (De Jode et al., 2019), although further research is necessary.
- (6)
Identifying the many missing CS would aid many areas in biology that rely on accurate species identification, such as assessment of ecosystem vulnerability or censuses of rare species.
- (7)
That CS are not a random sample of extant nominal species has two contrasting consequences: first, undiagnosed CS impact inferences from species data sets (e.g. presence, abundance, phylogeny) regarding ecological and evolutionary processes; and secondly, predictions of the presence of CS is theoretically possible, and could be used to focus future genetic studies on where CS are most likely to be discovered.
ACKNOWLEDGEMENTS
The authors declare that there are no conflicts of interest to disclose. We are grateful for help from T. Antonini, L. Barrau (literature reviews), C. Beaussi (library resources), J. Claude (APE package), A. Kroh (WoRMS), B. Tran (coding), and A. Baumel, A. Millon, J.-P. Féral, and J.-G. Harmelin (general advice). A. E. C. was funded by the DEVOTES (DEVelopment Of innovative Tools for understanding marine biodiversity and assessing good Environmental Status) project, funded by the European Union under the 7th Framework Programme, ‘The Ocean of Tomorrow’ Theme (grant agreement no. 308392), www.devotes-project.e.
Open Research
DATA AVAILABILITY STATEMENT
All codes and data files are available in a permanent public repository: https://osf.io/e4xm2/?view_only=0dd0a305584048a08cbdcff13b13d415.