Unveiling the complexity and ecological function of aquatic macrophyte–animal networks in coastal ecosystems
ABSTRACT
Network theory offers innovative tools to explore the complex ecological mechanisms regulating species associations and interactions. Although interest in ecological networks has grown steadily during the last two decades, the application of network approaches has been unequally distributed across different study systems: while some kinds of interactions (e.g. plant–pollinator and host–parasite) have been extensively investigated, others remain relatively unexplored. Among the latter, aquatic macrophyte–animal associations in coastal environments have been largely neglected, despite their major role in littoral ecosystems. The ubiquity of macrophyte systems, their accessibility and multi-faceted ecological, economical and societal importance make macrophyte–animal systems an ideal subject for ecological network science. In fact, macrophyte–animal networks offer an aquatic counterpart to terrestrial plant–animal networks. In this review, we show how the application of network analysis to aquatic macrophyte–animal associations has the potential to broaden our understanding of how coastal ecosystems function. Network analysis can also provide a key to understanding how such ecosystems will respond to on-going and future threats from anthropogenic disturbance and environmental change. For this, we: (i) identify key issues that have limited the application of network theory and modelling to aquatic animal–macrophyte associations; (ii) illustrate through examples based on empirical data how network analysis can offer new insights on the complexity and functioning of coastal ecosystems; and (iii) provide suggestions for how to design future studies and establish this new research line into network ecology.
I. INTRODUCTION
Network science offers innovative tools to explore the complex ecological mechanisms regulating species associations and interactions (Delmas et al., 2019). Rapid theoretical developments from multiple domains have recently provided researchers with new analytical and modelling tools, which allow us to venture beyond species-specific processes and to achieve a more realistic characterisation of the mechanisms regulating natural communities (Dunne, 2006; Landi et al., 2018; Delmas et al., 2019). This development has led to an increasing recognition that ecological interactions play a fundamental role in determining how ecological systems respond to environmental change (Woodward et al., 2010; Valiente-Banuet et al., 2015; Bruder et al., 2019). Several studies have revealed that the effects of direct drivers of species loss – such as global climate change or human overexploitation – can propagate through the many paths connecting species into complex ecological networks, causing secondary extinctions and even driving entire systems to collapse (Dunne & Williams, 2009; Strona & Lafferty, 2016; Strona & Bradshaw, 2018). This insight calls for increasing efforts to improve our understanding of the mechanisms controlling network dynamics and responses to perturbations which, in turn, requires the collection of proper data. Although the availability of information on the quality and intensity of ecological interactions is growing steadily (Poelen, Simons & Mungall, 2014), study effort is unequally distributed, hindering our ability to generalise and compare knowledge across different ecosystems.
Aside from food webs, which form an independent, long-standing research field studied through specific theoretical and computational tools (Dunne, 2006), there is a disproportionately large (and still growing) number of studies addressing plant–pollinator networks compared to other kinds of interaction networks (Fig. 1). Other ecological interactions that have been investigated using network analysis (Fig. 1) include those between hosts and parasites (Runghen et al., 2021), bacteria and phages (Weitz et al., 2013), plants and seed dispersers (Donatti et al., 2011), plants and herbivores (Araújo, 2016), plants and epiphytes (Naranjo et al., 2019), plants/animals and their microbiota (Bennett, Evans & Powell, 2019), ants and their partner organisms (Cagnolo & Tavella, 2015), and cleaners and their clients (Quimbayo et al., 2018). Such studies have provided important insights into the mechanisms regulating natural systems and advanced our understanding of the relationship between network structure and ecological stability (Bascompte & Jordano, 2007; Fontaine & Thébault, 2015; Welti, Helzer & Joern, 2017; Vizentin-Bugoni et al., 2018). However, to the best of our knowledge, no study has explicitly focused on the complexity of aquatic macrophyte–animal interaction networks. Here and throughout the text we refer to ‘macrophyte’ as a general term including both macroalgae and aquatic rooted vascular plants.
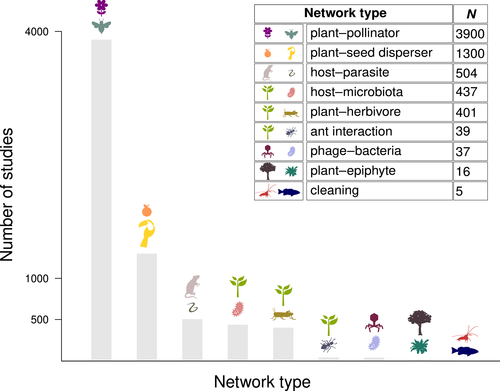
In this review, we investigate the reasons why macrophyte–animal systems have so far been largely neglected by network scientists. We then illustrate, aided by examples based on literature data, how networks analysis and modelling offer a powerful and flexible framework to explore a broad suite of fundamental ecological questions. Finally, we discuss practical ways to promote the establishment of this new field of research and its integration into coastal ecology. While our analysis mainly focuses on macrophyte–animal associations in the marine environment, we believe the approach proposed herein could provide novel insights into the functioning and ecological structure of freshwater and brackish ecosystems as well.
II. THE IMPORTANCE AND DIVERSITY OF MACROPHYTES IN COASTAL ECOSYSTEMS
Macrophytes dominate most freshwater and shallow marine littoral habitats (Murphy et al., 2019; Wernberg et al., 2019). In coastal waters, macroalgae occur in sparse to dense assemblages as well as in large underwater ‘forests’, with large brown algae (kelp) canopies alone occupying more than 25% of the world's coastal areas (Wernberg et al., 2019). Around 150000 km2 of seagrass meadows have been mapped globally (Green & Short, 2003; Duffy et al., 2019), while according to model estimates seagrass cover might extend to up to 1.6 million km2 (Jayathilake & Costello, 2018; Duffy et al., 2019). Hot spots of freshwater macrophyte diversity are found in the Neotropics (Murphy et al., 2019). In the marine realm, seagrasses are more diverse in the Indo-Pacific region (Green & Short, 2003), while macroalgal diversity exhibits no clear latitudinal pattern (Bolton, 1994). In relatively species-poor macrophyte systems, such as kelp forests or temperate seagrass beds, macrophyte genetic diversity plays a potentially important role for ecosystem functioning and stability (Reusch et al., 2005; Salo & Gustafsson, 2016; Wernberg et al., 2018). One peculiar system in this context is the Baltic Sea, a large brackish water body with a strong salinity gradient, which allows the coexistence of freshwater and marine macrophyte species, resulting in high macrophyte diversity (Hällfors & Niemi, 1981).
Macroalgae and seagrasses play pivotal roles in coastal ecosystems. As prolific primary producers, they constitute key basal sources of energy in littoral food webs, which they fuel with both fresh vegetal material and detritus (Christie, Norderhaug & Fredriksen, 2009). Besides their trophic role, macrophytes participate in a broad range of other ecological associations, hence promoting animal diversity. By their mere presence, they generate complex three-dimensional structures, which provide critical habitat for a variety of organisms with various requirements and life strategies. Organisms benefitting from such structures range from epiphytic algae to invertebrates and fish seeking shelter from predators or hydrodynamics, or a suitable substrate on which to nest or breed (Christie et al., 2009; Duffy et al., 2019). Furthermore, macroalgae and seagrasses contribute to multiple ecosystem services: by providing essential habitats and foraging and nursery grounds for many fish species of commercial interest, they sustain coastal fisheries globally (Nordlund et al., 2018). Macrophytes also mitigate pollution and eutrophication, which represent some of the main threats to the integrity of coastal ecosystems (Howarth et al., 2000). As a result of their metabolism, macrophytes can ‘filter’ water, retaining nutrients in their biomass and reoxygenating the sediments (McGlathery, Sundbäck & Anderson, 2007). By absorbing carbon dioxide from the water column and sequestrating it in the seafloor they act as important carbon sinks in coastal waters (Duarte et al., 2010; Krause-Jensen & Duarte, 2016; Watanabe et al., 2020). Lastly, they provide physical protection for coastlines and mitigate beach erosion – services which are imperilled by progressing, climate-induced change (Ondiviela et al., 2014; Innocenti, Feagin & Huff, 2018).
III. WHY DO WE NEED MACROPHYTE–ANIMAL NETWORKS?
(1) Macrophyte communities in a changing world
Macrophyte communities are exposed to multiple local and global stressors, including nutrient enrichment, pollution, outbreaks of grazers caused by overfishing of predatory fish, invasive species, increasing sedimentation and temperature rise (Strain et al., 2014; Takolander, Cabeza & Leskinen, 2017). These major disturbance factors are driving extensive regime shifts in seaweed and seagrass assemblages worldwide, often resulting in the loss of complex, persistent canopy-forming macrophytes in favour of simpler, transient mat-forming species with lower ecological value (Benedetti-Cecchi et al., 2001; Airoldi et al., 2014; Mineur et al., 2015; Unsworth et al., 2015; Gorman et al., 2020).
Changes in macrophyte community composition inevitably alter the range of ecological services that they provide to their associated fauna via trophic and non-trophic relationships, with consequent shifts in animal diversity and abundance (Benedetti-Cecchi et al., 2001; Edgar et al., 2004; Lilley & Schiel, 2006). The mechanisms through which disturbance propagates from macrophyte to animal communities are complex and elusive: as macrophyte–animal systems are embedded in larger littoral food webs, perturbations at the level of macrophyte–animal links are likely to propagate to higher trophic levels through cascading effects (Cordone et al., 2018; Strona & Bradshaw, 2018) and may affect species of particular ecological and/or commercial value. In worst-case scenarios, these domino effects might ultimately drive whole regime shifts in coastal ecosystems (Wernberg et al., 2016).
The primary role of macrophytes in maintaining littoral biodiversity and coastal ecosystem functioning, and their vulnerability to human activities and climate change, call for a deeper understanding of the complex mechanisms through which macroalgae and seagrasses sustain their associated biological communities. In this context, ecological network analysis might provide a powerful tool to investigate the structure and dynamics of macrophyte–animal associations.
(2) Macrophyte–animal networks: reinventing the wheel?
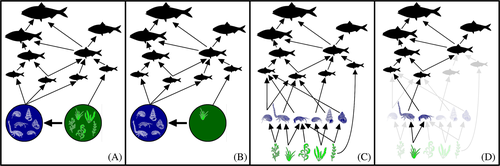
Studies of aquatic macrophytes (and their associated fauna) have largely focused on coastal food webs. These are typically represented as a set of nodes, symbolising different taxa or functional groups, and links, illustrating the possible trophic interactions between each given pair of nodes (Fig. 2). Why are we then claiming novelty in the application of network theory to macrophyte–animal associations? To answer this question, it is important to clarify that when discussing macrophyte–animal networks, we are not referring to a subset of nodes and links extracted from a complete coastal food web but, instead, to an organic entity that stands on its own.
Considerable work has been done to clarify the trophic relationships between aquatic macrophytes and their consumers, especially through stable isotope analysis (Crawley et al., 2009; Porri, Hill & McQuaid, 2011; Bessa, Baeta & Marques, 2014; Kahma et al., 2021; Liénart et al., 2021). However, when considered as parts of ‘complete’ coastal food webs, the macrophyte–animal component is often regarded as a generic source of energy for the upper trophic levels, while the dynamics of the links directly connecting macrophytes to animal species are often neglected. With few exceptions (Coll et al., 2011; Kéfi et al., 2015; Cordone et al., 2018; Momo et al., 2020), macrophyte and invertebrate links are frequently pooled into single nodes in the food-web representation, while only links connecting nodes at higher trophic levels are represented in full detail. This imbalance in detail is mostly due to the fact that information on trophic interactions in coastal food webs frequently stems from the analysis of predators' diets (Olivier et al., 2019), while identifying large numbers of small-body-sized organisms – and the trophic interactions they participate in – is more challenging.
However, this over-simplification is likely to affect our understanding of the processes taking place at the macrophyte–animal interface. In particular, combining multiple species into single food-web nodes fuels the assumption that pooled species are ecologically equivalent, and hence that changes in their diversity or relative abundances will have no effects on food-web functioning and other ecosystem processes (Fig. 2A, B). This perception stands in sharp contrast with various empirical and observational studies showing how changes in macrophyte community composition can have strong effects at higher trophic levels (Edgar et al., 2004; Wernberg et al., 2016). Conversely, identifying the potentially complex structure of macrophyte–animal associations might reveal novel patterns, and enhance our ability to model processes at the macrophyte–animal level and their potential effects on the rest of the food web (Fig. 2C, D).
- As a general rule, in food webs, each node can simultaneously be a consumer or a resource for other consumers. This makes them unipartite, in contrast with other types of ecological networks (such as mutualistic and host–parasite networks), where nodes can often be categorised into distinct groups (e.g. plants or pollinators), with links occurring only between (but not within) groups. When such networks include two categories, they are bipartite, and when they include more types, they are multipartite. This distinction between uni-, bi- and multipartite networks implies differences in how they are studied and in how their structural properties are interpreted in ecological terms (Dormann et al., 2017). Technically, a unipartite food web is an ecological network. However, for simplicity, in the following we will refer to food webs in their classical, trophic sense as ‘food webs’ and to all other kinds of bipartite networks (such as those in Fig. 1) as ‘ecological networks’. The object of this review will be animal–macrophyte bipartite networks, as distinct from unipartite coastal food webs.
- We look at generic associations (i.e. not only at trophic interactions, as in the case of food webs). In aquatic macrophyte–animal systems there are multiple types of potential associations, each possibly revealing different forms of interactions including herbivory, habitat dependency and various form of symbiosis at different positions in the mutualism–parasitism continuum (Leung & Poulin, 2008). Such interactions are not equivalent from an ecological perspective and might potentially be used to identify distinct ecological networks (see Section V). Likewise, similar patterns of associations can emerge from different types of interactions. However, while detecting spatial associations can be relatively easy, detecting interactions is challenging, especially in aquatic ecosystems. For this reason, throughout this review, we use the generic term ‘association’ to refer to animal species that are preferentially or exclusively found on or in close proximity to a macrophyte. Our underlying assumption is that the recurring presence of an animal species on a given macrophyte species points towards the existence of a certain degree of dependency, prompting the expectation that the target animal would disappear (or substantially drop in abundance) following the loss of the host macrophyte.
- We are interested in species-specific associations; that is, we do not consider (a priori) macrophyte or animal species as interchangeable nodes in a network. Instead, we consider network analysis as an important tool to reveal to what extent species loss and changes in species composition can modify the overall structure of association networks, thereby potentially modulating their persistence – with subsequent effects on other species with direct links or indirect paths to species lost from or acquired by the community.
IV. ARE MACROPHYTE–ANIMAL ASSOCIATIONS STRUCTURED?
Despite a lack of focused research on the structure of macrophyte–animal networks, several studies have explored different aspects of macrophyte–animal associations, both in the freshwater (see Gerrish & Bristow, 1979; Biggs & Malthus, 1982; Rooke, 1984; Kurashov et al., 1996; Mhlanga & Siziba, 2006) and marine realms (e.g. Virnstein & Howard, 1987; Dhargalkar, Burton & Kirkwood, 1988; Russo, 1990; Parker, Duffy & Orth, 2001; Trowbridge, 2004; Pereira et al., 2006; Huang et al., 2007; Wikström & Kautsky, 2007; Gustafsson & Boström, 2009; Nakamoto et al., 2018). Studies to date span from polar (e.g. Amsler et al., 2015) to temperate (e.g. Piazzi et al., 2018) and tropical (e.g. Tano et al., 2016) waters. As we demonstrate in Section VI, these studies offer candidate material to build and explore proof-of-concept macrophyte–animal networks. However, only a small fraction of published literature/data sets includes the kind of information needed to compile a network. In particular, many available studies focus on the faunal assemblages of a few selected macroalgae and seagrass species (Russo, 1990; Gee & Warwick, 1994; Lippert et al., 2001; Parker et al., 2001; Chemello & Milazzo, 2002; Saarinen, Salovius-Laurén & Mattila, 2018), while there is comparatively less material providing suitable data to build detailed ecological networks.
Beyond the taxonomic challenges mentioned above, which can now be tackled with next-generation molecular techniques (Leray & Knowlton, 2015), there are other practical aspects which complicate a comprehensive collection of macrophyte–animal associations, for example, related to sampling (Jordano, 2016a). However, in addition to these, more conceptual reasons might lie behind the tendency for ecologists to look at pairwise associations between macrophytes and animals without attempting to combine them into ecological networks. Most notably, there is a common perception that, unlike their terrestrial analogues, aquatic macrophyte–animal systems are permeated by widespread generalism in resource–consumer associations (Taylor & Cole, 1994; Bates & DeWreede, 2007). If this view was true, it would weaken the expectation for structural patterns and make a network approach ineffective and no more informative than, for instance, quantitative studies using plant biomass and diversity as predictors of animal abundance and diversity (Kraufvelin & Salovius, 2004; Gollety, Thiebaut & Davoult, 2011; Zhang et al., 2021).
Often, in the aquatic literature, animals found associated with only a few species of macrophytes are classified as ‘generalists’ (Lewis, 1987; Taylor & Cole, 1994; Lippert et al., 2001). The tendency to consider specialisation in macrophyte–animal systems as a binary property (where only animals associated with a single macrophyte species are considered ‘specialists’) might have inflated the idea of macrophyte–animal interactions as systems dominated by generalists (Devictor et al., 2010). Such a perception might have been exacerbated by the tendency to focus on dominant macrophyte (or associated animal) species (Russo, 1989; Taylor & Cole, 1994) and hence to overlook potential patterns of specialised associations involving less-abundant species (Arrontes, 1999). Taken together, these aspects might have discouraged attempts to explore macrophyte–animal associations through network analysis, by conveying the biased impression of a lack of ecological structure.
By contrast, as our theoretical understanding of the ubiquitous importance of ecological interactions (and their complex organisation) has improved (Jordano, 2016b), it has also become clearer that such a view might represent a severe oversimplification. This is further supported by various observational and experimental studies on macrophyte–animal systems which have demonstrated that there actually is structure in these associations.
Among the variety of factors that shape animal assemblages in submerged vegetated habitats, macrophyte morphological complexity and architecture (Gee & Warwick, 1994; Chemello & Milazzo, 2002; Huang et al., 2007) seem to play an important role in driving host selection by fauna. Adding secondarily to such selection is the chemical composition – and hence often nutritional quality – of macrophytes (Hay et al., 1987; Hay, Duffy & Fenical, 1990; Duffy & Hay, 1991; Lastra et al., 2008). Although this rarely results in tight specialisation (i.e. animals tend not to depend on single macrophyte species), evidence of different degrees of dietary or habitat specialisation does exist (Trowbridge, 1993, 2004; Sotka, 2005; Baumgartner, Pavia & Toth, 2014).
While a few studies seem to support the idea that animals and macrophytes interact randomly (Bates & DeWreede, 2007; Amsler et al., 2015), there is growing evidence that the composition, diversity and relative abundance of faunal assemblages vary substantially (and in a non-random fashion) in response to changes in associated macrophyte communities (Lippert et al., 2001; Chemello & Milazzo, 2002; Bates, 2009; Schaal et al., 2016; Machado et al., 2019). Furthermore, experiments involving canopy removal have shown how the loss of macroalgal canopy and the subsequent invasion by algal turfs deeply changes the composition and structure of understorey invertebrate assemblages (Dayton, 1975; Benedetti-Cecchi et al., 2001, 2015; Strain et al., 2014; Rindi et al., 2017).
Overall, these studies provide strong support for the idea that, rather than being governed by randomness, macrophyte–animal associations might instead be structured. This, in turn, implies that ecological network analysis can provide useful theoretical and analytical tools to explore macrophyte–animal associations and unveil their organisation. In the following sections, we dig deeper into the variety of ecological interactions involved in macrophyte–animal systems and the challenges they pose for network analysis. Through practical examples and different approaches, we then illustrate the wide range of information that one can obtain by assembling ecological networks of macrophytes and animals, and the benefits that such an innovative approach can bring to this field of study.
V. POTENTIAL CHALLENGES AND PITFALLS IN ASSEMBLING MACROPHYTE–ANIMAL NETWORKS
(1) The complexity of ecological interactions in macrophyte–animal systems
The ecological relevance of macrophytes in coastal communities is not limited to their trophic role, but encompasses a complex range of non-trophic antagonistic, mutualistic, and commensal relationships (York et al., 2018; Momo et al., 2020; see also Section II). Sometimes the boundaries between interaction types are blurred and assessing whether an animal species is beneficial or detrimental to the associated macrophytes can be complicated (see Section V.2). For example, a common form of interaction in macrophyte systems is epiphytism. In general, macroalgae and seagrasses facilitate sessile invertebrates by providing surfaces for attachment, and hence decreasing competition for space (York et al., 2018). By growing on kelp fronds, colonial hydrozoans reduce competition for space in the benthos and increase their accessibility to food resources in the water column. At the same time, they provide their macrophyte hosts with ammonium, while defending them from grazers (Hepburn & Hurd, 2005; González-Duarte, Megina & Subida, 2020). However, epiphytic organisms, such as bryozoans, often have negative effects on macrophytes, as they reduce access to light and diffusion of nutrients and gases (Muñoz, Cancino & Molina, 1991; Hurd, Durante & Harrison, 2000).
Other mutualistic interactions can be observed between mesograzers, such as amphipods, and macroalgae. Amphipods facilitate macroalgae by feeding on smaller epiphytic algae that compete for light and space, while macroalgae provide amphipods with a refuge from predators (Amsler, McClintock & Baker, 2014).
Another aspect that can complicate interpreting the nature of ecological interactions in macrophyte–animal systems is that this nature might shift when abiotic conditions change. For example, sponges and seagrasses can establish commensalistic interactions, where the growth of the former has a neutral effect on the latter (Archer, Hensel & Layman, 2018). However, under high nutrient loading, the interaction can become detrimental for the seagrass and shift to antagonistic (Archer et al., 2018).
In specific settings, macrophytes too might have a negative impact on the associated fauna, as in the case of seaweeds with large fronds, which can impede faunal settlement and grazing through the so-called ‘whiplash effect’ (Iken, 2012). Periodic shedding of outer cell walls by some algae also removes epiphytic layers and associated grazers (Russell & Veltkamp, 1984). However, in general, canopy-forming macroalgae maintain diversified assemblages of understorey algae and invertebrates (Benedetti-Cecchi et al., 2001; Strain et al., 2014).
The general picture emerging from these examples of interactions is composite and suggests that, to disentangle the complexity of macrophyte–animal associations and to explore their structure effectively, we need an appropriate analytical approach.
(2) A multi-layer framework
How to synthesise the complexity of species interactions within a single theoretical and analytical framework is a significant ecological challenge (Jordano, 2016b). For instance, in terrestrial plant–animal systems, many different trophic (e.g. plant–herbivores) and non-trophic interactions (e.g. plant–pollinators and plant–seed dispersers) have been shown to play simultaneous, intertwined roles in ecological processes (Pocock, Evans & Memmott, 2012). Traditionally, different kinds of interactions have been addressed as separate entities in specific branches of network ecology (Bascompte & Jordano, 2007; Donatti et al., 2011; Araújo, 2016). Recently, attempts have been made to move beyond the strict categorisation of networks and to investigate multiple types of ecological interactions within a unifying, multi-layer network framework (see Melián et al., 2009 and Pocock et al., 2012 for plant–animal systems, and Lafferty et al., 2008 and Kéfi et al., 2015 for food webs). Networks composed of multiple layers of interactions might offer an increasingly realistic representation of ecological complexity (García-Callejas, Molowny-Horas & Araújo, 2018) by capturing better the multifaceted nature of species interactions in natural systems.
Given the composite suite of relationships linking macrophytes and their associated fauna, a multi-layer network approach appears an optimal choice to explore these systems. However, while the approach has solid theoretical foundations from physics (see Pilosof et al., 2017), integrating multiple interaction types into single networks poses many practical challenges. As discussed above, identifying which interactions lie behind observed associations is, in general, extremely challenging. This is true also for well-investigated systems: for example, ‘cheating’ strategies in plant–pollinator (Thakar et al., 2003) or client–cleaner interactions (Guimarães Jr et al., 2007) greatly complicate the distinction between mutualistic and antagonistic interactions. In aquatic macrophyte–animal systems, the simultaneous sampling (and identification) of multiple kinds of interactions is an ambitious task, due to the variability of the associations in space and time (Christie et al., 2009; Machado et al., 2019). Moreover, there are practical challenges in identifying and recording species interactions in the marine environment compared to terrestrial habitats. However, the promising results obtained by the first studies on multi-layer networks (Lafferty et al., 2008; Melián et al., 2009; Pocock et al., 2012; Kéfi et al., 2015) should encourage ecologists to embrace the challenge.
One practical way to assemble multiple-interactions networks is to take advantage of pre-existing knowledge on species interactions within a target area or system (Kéfi et al., 2015; Puche et al., 2020). As an example, Kéfi et al. (2015) assembled a comprehensive network of trophic and non-trophic interactions in the Chilean rocky shore intertidal community from a large data set encompassing studies conducted over 40 years. However, such an approach is an exception rather than a rule. In the specific case of macrophyte–animal associations, information on the nature of ecological interactions is sparse. Although this has been thoroughly investigated in some systems, as is the case for Antarctic seaweeds and their associated fauna (Amsler et al., 2014; Heiser et al., 2020; Momo et al., 2020), sufficient pre-existing information is typically lacking. This knowledge gap highlights the need for de-novo observational and experimental studies. Considering the motivation behind this review, we are certainly at too early a stage to set such an ambitious goal, and it therefore seems appropriate to start by looking at single layers of associations. Nevertheless, it is important to identify a multi-layer representation of macrophyte–animal networks as the long-term target, to ensure that the steps taken now will continue to move us in the right direction.
In the following section, we attempt to start that process by reconstructing some preliminary macrophyte–animal networks based on data sets available in the literature. In line with the considerations above, our source data do not include details about the type of interactions (e.g. mutualism, commensalism, antagonism) behind the observed associations. However, they allow us to illustrate how a network perspective can be used to extrapolate novel valuable information not attainable with other approaches. They also allow us to demonstrate how a network approach can significantly change our understanding of the macrophyte–animal systems under study. Our hope is that these simple examples will stimulate the interest of researchers to either reconsider already-collected data from a new perspective and/or to start collecting new data with network analysis in mind.
VI. INVESTIGATING MACROPHYTE–ANIMAL ASSOCIATIONS THROUGH NETWORK ANALYSIS: EXAMPLES OF APPLICATIONS
In this section, we use empirical data to reconstruct a suite of bipartite macrophyte–animal networks, investigate their structure and properties and model the effects of disturbance. The data sets were selected based on their suitability for network analysis and not following any specific ecological or geographical criteria. All analyses were run in the free software Python 3.6.9 and R 3.6.3 and the code allowing full replication of the analyses is freely available at https://github.com/mancfede/macrophyte_animal_networks.
(1) Building networks from literature data
We identify two categories of data sets that can be used to build tentative ecological networks of macrophytes and animals. The first consists of potential interaction data, which are most often represented by checklists of the fauna associated with a given and often small set of macrophyte species. Usually, such data sets report quantitative information (e.g. the number of individuals or the biomass of a given animal species found on a given macrophyte individual/species). The second category consists of co-occurrence data, for example of lists of macrophyte and animal species found within the same sampling unit. With appropriate caveats (Freilich et al., 2018; Blanchet, Cazelles & Gravel, 2020), this information can be used to identify pairwise associations, by establishing which species tend to be found together more often than predicted by chance and/or by a set of environmental/ecological factors. Pairwise associations can then be combined into association networks (Araújo et al., 2011; Berry & Widder, 2014; Harris, 2016). It should be highlighted that the idea of using co-occurrence to infer ecological interactions is currently debated (Freilich et al., 2018; Blanchet et al., 2020). A key aim of this review is to encourage studies specifically designed to collect reliable data on species interactions. Thus, we present here the approach that is best able to obtain additional information from data originally collected for purposes different from network analysis. The distinction between the two categories can be subtle, as data of fauna associated with macrophytes are somehow a (stricter) form of co-occurrence. However, the following two examples may act to clarify the distinction.
As an example of the first category (data set of pairwise associations/checklists of animals associated with a given macrophyte individual/species), we use the data from Table 1 in Huang et al. (2007). This table reports the number of gammaridean amphipods found on eight different dominant macroalgal species from the West Antarctic Peninsula. Ideally, this measure could be used to build a weighted network. The most straightforward way to do this would be to generate a link between an algal and an amphipod species, with the weight of the link equalling the number of individuals reported in the table. A further step would be to standardise the link weights by dividing them by the total number of amphipod individuals (of all species). Such a choice would emphasise the absolute strength of the interactions, by giving higher weights to dominant species compared to rare ones (Fig. 3A). An alternative approach to standardise weights would be to divide the number of amphipods found on given algae by the total number of amphipods of that species. Such an approach would emphasise patterns of amphipod specialisation, without penalising rare species occurring at low abundances (Fig. 3B).
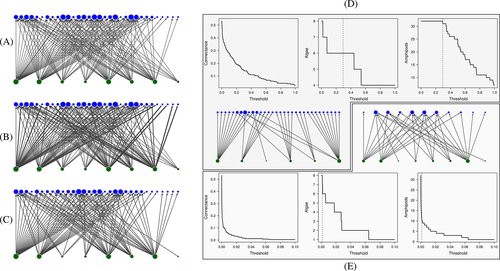
Regardless of the criterion adopted to attribute and standardise weights, a weighted network can be investigated without transformation, or first be transformed into an unweighted network. Such transformation requires identifying a threshold for the interaction strength below which links are disregarded. For example, referring again to the data set from Huang et al. (2007) one could build an unweighted network (Fig. 3C) by arbitrarily considering an amphipod as associated with a target alga if at least 1% of the individuals of a given amphipod species was found on the target alga. This is equivalent to imposing a threshold for interaction strength of 0.01 on the network reported in Fig. 3C. While the strongest potential associations can be derived directly from the original table, exploring how the density of ecological interactions declines as we raise the threshold for interaction strength could yield insights into the degree of specialisation of the system.
Although setting a threshold might be arbitrary in itself, the exploration of how different thresholds affect the outcome can be used to guide the choice. For example, in the data set from Huang et al. (2007), consider the weighted network obtained by dividing the number of individual amphipods found on one alga by the total number of individuals of the same amphipod species found on all algae (Fig. 3B). In describing relevant amphipod/algae patterns of specialisation regardless of species abundance, it might make sense to use the minimum value ensuring that all the amphipod species are included in the network. This threshold turned out to be 0.295 (Fig. 3D). However, in the case of the network with weights standardised by the total number of individuals (Fig. 3A), it might make sense to choose a threshold regardless of its effect on the number of amphipod species left in the network. Since one interesting aspect of that network would be identifying strong associations possibly having important ecosystem effects, choosing a threshold which simultaneously eliminates links involving few individuals and rare amphipod species could be a good choice. A possible option could be that of selecting as a threshold the minimum interaction strength ensuring that all algae are left in the data set (i.e. 0.0014) (Fig. 3E). This threshold leaves us with 11 amphipod species (from the initial 32), and a quite simplified version of the network possibly representative of the ‘core’ interaction set. However, there is no right or wrong criterion in this regard, nor a ‘golden rule’ applicable to all situations.
As an example of the second possible category of data sets that might be used to obtain a network (that is data sets reporting the independent occurrence of macrophytes and animals across a set of samples), we compiled two macrophyte–animal networks using a data set from Nakamoto et al. (2018). This data set was generated by assessing the diversity and biomass of macrophytes in a subtropical seagrass–seaweed mixed bed in Okinawa. While the original study aimed to assess the effect of macrophyte phylogenetic diversity on the species diversity of associated epibenthic invertebrates, the same data can be used to identify, with a network approach, relevant associational patterns. As examples, one may score which macrophyte–invertebrate associations are maintained over time and the degree of specialisation of the system.
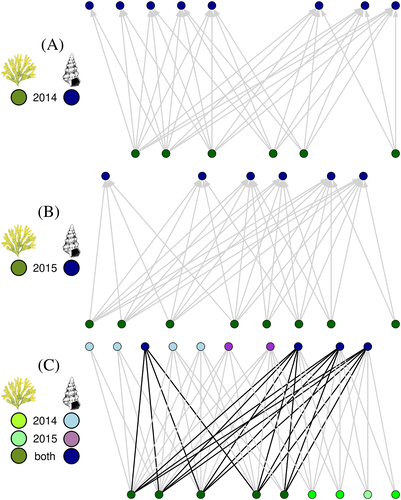
In the original study, researchers set twelve 50 cm × 50 cm quadrats and then counted mobile epibenthic invertebrates in each quadrat in two consecutive years. Due to the sampling procedure, the data set provides information only on the number and identity of invertebrates and the respective biomass of all macrophytes found within the same quadrat. This does not permit a direct network reconstruction. We therefore inferred potential associations based on the co-occurrence between animal and macrophytes across quadrats. For this, we first quantified the observed co-occurrence for any possible macrophyte–animal pair as the total number of quadrats where both candidate interacting partners were found. Then, to determine which species associations occur more frequently than expected by chance, we compared the observed co-occurrence with that measured in 1000 randomised assemblages obtained by randomly reallocating animals across quadrats. These assemblages were constructed by keeping the number of occurrences per macrophyte and animal operational taxonomic unit (OTU) fixed, while permitting the number of occurrences per quadrat to vary freely. In particular, we computed, for each candidate pair, a P-value as the fraction of randomised assemblages where the target pair had a co-occurrence equal or higher than that observed in the real assemblage. Finally, we generated a network by collating all the pairs where P < 0.05. We replicated the procedure individually for samples collected either in the first (2014) or second (2015) year of the survey, hence obtaining two distinct networks (Fig. 4A, B). These two networks can optionally be combined into a multitemporal network, thus allowing separation between species associations present during both years of sampling, and those present in an individual year only (Fig. 4C).
(2) Exploring ecological network structure
To illustrate the wider utility of network analysis for distinguishing differences versus similarities in the constellation of biotic associations over time, consider the two networks in Fig. 4A, B. Such networks will ideally represent the same system at two different times. We can look at those networks from a qualitative perspective, focusing on the taxonomic identity of the nodes involved in the links. In this way, the two networks can be compared in a straightforward manner, highlighting common elements and differences in the association set. The core structure of the networks appears stable between years, with more than 30% of the links being shared between the two networks regardless of changes in species composition (Fig. 4C), and all the links preserved when focusing only on species present in both surveys. These results provide support for the idea that the associations between macrophytes and animals indeed might be structured and hence be particularly suitable for network analysis. But what if, instead, we had found profound differences between the link identities? Would that have meant that the two networks were completely different?
This is where the power of network analysis resides. In Fig. 4A, B, we have plotted species so that their horizontal position is kept consistent in the different networks in order to facilitate comparison. Yet, this is the only aspect that hints at species' identity. Other than that, all the species are represented in the network with the same symbol. On one hand, this is a graphical choice: we might have used different silhouettes for the different species, or different node colours, or spelt out species' names. On the other hand, the main value of using networks is that of having an abstract, simplified and yet extremely informative representation of a complex system. In such a representation, the taxonomic identity of nodes becomes irrelevant, while what really matters is how those nodes are arranged and how that arrangement generates global and local structural patterns. In this way, networks offer a convenient framework to obtain objective measures useful to describe and compare natural systems, even when the respective networks are inhabited by taxonomically fully distinct assemblages.
Several metrics have been developed for characterising network structure. The main distinction is between local measures (i.e. properties of a node or link) and global measures (i.e. properties of the whole network). Additionally, there are ‘meso-structure’ indicators, such as core–periphery metrics. It should be noted that there are differences between the structural metrics typical of unipartite networks, such as food webs (Dunne, 2006), and those typical of bipartite networks (Ings et al., 2009) such as plant–pollinator networks and macrophyte–animal networks as we are presenting them here. In the following examples, we will refer mostly to metrics for bipartite networks. To illustrate how they may be used to resolve key features of macrophyte–animal networks, we will next consider different descriptors of specialisation and of the importance of different nodes.
To characterise network structure, both local and global measures can be used in ecological analysis. The most obvious example of a local measure is node degree, which is the number of in- and out-coming links of a target node. This local measure can be used to obtain a global ‘fingerprint’ of the network, which is the probability distribution of node degrees (a network's degree distribution). A typical procedure in network science is plotting the degree distribution with both axes (x = node degree; y = frequency) on a log scale (Fig. 5B). Such analysis can provide an immediate visual representation of specialisation patterns.

The network in Fig. 5A, which we reconstructed from the data obtained in Table 2 from Chemello & Milazzo (2002), is one of the largest networks we were able to assemble. It includes six macrophyte species, 57 invertebrate (mollusc) species and 159 links. As the information reported in the study refers to three replicate samplings, each link in our network represents a potential association identified in all three replicates. As shown in Fig. 5B, the degree distribution of the invertebrate species follows a power law, where most invertebrate nodes tend to have few connections (1 link), while only a few nodes show a relatively high generalism (>5 links).
Less-obvious measures attribute importance to a node based on different criteria which consider broader portions of the network. Ideally one could obtain some measure of node importance and then compare it with other species features to test various hypotheses. For example, one could test whether the position of a node in a network is affected by specific ecological traits and, in turn, how such network position could affect the vulnerability of the target species to indirect effects of environmental change propagating through network links (Strona & Lafferty, 2016; Escribano-Avila et al., 2018).
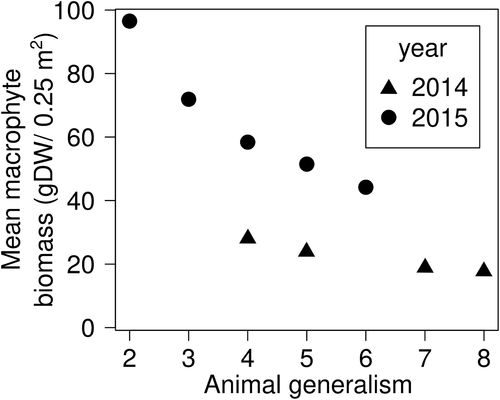
As an example, we compared the specialisation of associations versus macrophyte biomass in the networks obtained from co-occurrence in the data from Nakamoto et al. (2018). As a general expectation, specialised consumers should rely on ‘stable’ resources (e.g. resources that are broadly distributed, locally abundant and available through time), providing a key to the robustness and stability of ecological networks (Strona & Lafferty, 2016). For this, we find support in the network observed by Nakamoto et al. (2018). In Fig. 6 we show that the more ‘specialised’ invertebrates (that is, those connected to fewer macrophytes in the network) are preferentially associated with macrophytes having the largest overall biomass. In theory, this could make networks more robust to oscillation in resource populations compared to cases where no relationship exists between consumers' specialisation and resource availability (Strona & Lafferty, 2016). Of course, an obvious question is whether the observed pattern reflects the actual specialisation in the animal community or, instead, the observed ‘specialisation’ is an artefact of macrophyte abundances. This is where network analysis becomes important. Although we expect to observe more animal species on the more common/abundant macrophyte species simply due to chance, the simultaneous consideration of the whole network allows us to assess whether the observed degree of specialisation and network structure can be explained by the sampling process alone or whether it results from non-random associations. The consistency between network links across the two sampling seasons, and the consistency of the trajectories observed in Fig. 6, seem to suggest that the pattern is indeed a product of network structure and specialisation of associations, and not a mere result of chance.
The structure of an ecological network can reveal different processes. For example, the tendency for sharing partners in mutualistic networks (nestedness) can promote diversity by attenuating competition for resources (Bastolla et al., 2009). Similarly, the organisation of an ecological network into fairly isolated clusters of highly connected interacting species can have important implications for network persistence/stability (Thébault & Fontaine, 2010). We explored nestedness in the network from Chemello & Milazzo (2002) reported in Fig. 5A (Fig. 7). In particular, we used the popular NODF metric (Almeida-Neto et al., 2008). The metric ranges from 0 to 100, with 100 being the score of a perfectly nested matrix. We compared the NODF value observed in the original network with the NODF values of 1000 null networks. We generated the null networks using an algorithm that randomises the position of animal–macrophyte links in the network without altering the total number of animals associated with a given plant. We then obtained a P-value as the fraction of null networks with a NODF value higher than that of the original matrix, and an effect size (Z) as [NODFobs–mean(NODFnull)]/std(NODFnull), where NODFobs is the observed NODF, and NODFnull is the set of NODF values measured in the null networks. We performed the analyses using the freely available NeD software (Strona et al., 2014).
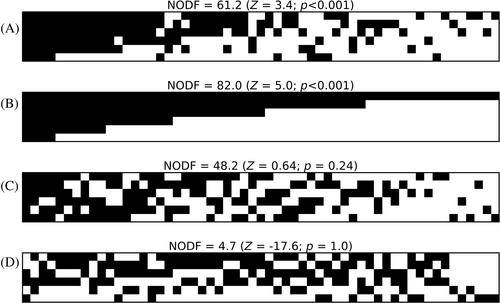
In addition to this analysis, for comparative purposes, we generated the ideally most and least nested networks. These can be obtained by randomly reallocating links in the network with the only constraints being maintaining the initial number of links, and at least one link per species in the network (i.e. avoiding the generation of empty rows and columns in the matrix representing the network). Alternatively, one can generate a totally random matrix (with the same constraints). In Fig. 7 we provide visual representations of the matrices corresponding to the original network (Fig. 7A), the most nested realisation (Fig. 7B), one random realisation (Fig. 7C), and the least nested realisation (Fig. 7D), together with their NODF, Z and P values. The original matrix (Fig. 7A) proved significantly nested (as commonly observed in ecological networks), and much closer to the ideal perfectly nested configuration (Fig. 7B) than to the random one (Fig. 7C), and very far from the least nested possible configuration (Fig. 7D). The presence of a nested structure is consistent with the expectation for ecological networks, because nestedness is an expected by-product of the same mechanisms illustrated above about consumers specialising on dependable resources (Strona et al., 2013). Furthermore, nestedness might be key to the emergence of complexity in multi-species communities, as the tendency to share interacting partners or resources can counteract the negative effects of interspecific competition and promote the coexistence of species (Bascompte & Jordano, 2006). Thus, the observed patterns provide circumstantial evidence confirming that the observed associations (and hence the higher-level structure emerging from those) are not a product of chance. Instead, they might indeed reflect ecological and co-evolutionary processes, although the purely illustrative nature of the example prevents us from hypothesising too far.
(3) Exploring the effect of disturbance on macrophyte–animal network structure
Networks offer a convenient framework to explore how different types of disturbance (e.g. environmental change) can propagate through (and possibly be amplified by) ecological interaction links and, more broadly, affect the structure and stability of the biological systems under study. In this section we provide two examples, one theoretical and one based on empirical data from a perturbation experiment. We then discuss the ecological information attainable by analysing macrophyte–animal network structure under different scenarios of disturbance.
(a) Mapping hypothetical co-extinction scenarios of macrophytes and animals
An informative exercise in ecological network analysis is exploring the robustness of networks to incremental species loss, that is to the subsequent removal of nodes (Memmott, Waser & Price, 2004; Dunne & Williams, 2009). The number of nodes to be removed, as well as the sequence of removal, depends on the specific objectives of the simulation. Similarly, the rules determining the effect of the loss of nodes and links (i.e. species and interactions) on the network can vary on a per case basis, depending on the starting hypotheses, the objectives of the analysis, the kind of network under examination, and the informative content of the network. For example, the rules that can be set for a weighted network differ from those for an unweighted network. The same applies to directed versus undirected networks and unipartite versus bipartite networks. In one of the simplest possible models, one can reiterate the procedure of removing one node at random from a network, and then remove the nodes left with no resources after the first removal, until no nodes are left in the network. In a food web, one can remove species in random sequence and prune the network from the portions left isolated from basal resources after each removal, hence simulating ‘extinction cascades’ (Dunne & Williams, 2009). In a bipartite network, such as a plant–pollinator network, it is appropriate to focus on the response of one of the two groups to the loss of interacting partners from the other group. For instance, a typical simulation consists of progressively removing plants, keeping track of the resulting pollinator diversity (or vice versa) (Memmott et al., 2004). The results can be plotted on a graph showing the decline in pollinator diversity following the removal of plant species (i.e. in a plot showing ‘primary extinctions’ versus ‘secondary extinctions’ or ‘co-extinctions’).
The removal of species could be based on some informed criteria, for example by combining data on species thermal tolerance with projections of future climate change (Strona & Bradshaw, 2018), or could be defined according to arbitrary criteria, such as random node removal. In the latter, the true value of the analysis is that of offering a way to compare the ‘intrinsic’ robustness of a network to disturbances. To demonstrate such an approach, we ran a set of co-extinction simulations (Fig. 8) in the network reconstructed from the data of Chemello & Milazzo (2002) (Fig. 5A), which maps 159 associations between six macroalgae and 57 animal species. In particular, we performed three sets of node-removal simulations (with 100 replicates per set) aimed at approximating worst, random and best scenarios of network collapse following macroalgal loss. For this purpose, we progressively removed macroalgae from the network in random order, or by choosing, at each step, either the most (in the worst-case scenario) or the least (in the best-case scenario) connected macroalgal species. The worst and best-case scenarios were simulated using a ‘greedy’ algorithm, that recalculates the number of connections per node at each step (as in Allesina & Pascual, 2009). After each macroalgal removal, we computed the relative fraction of animal species which were still connected to algae. We finally averaged the results, for each scenario of node removal, across the 100 replicates. For comparison, we replicated the same exercise in one of the most complete (and most famous) plant–pollinator (or better, plant–visitor) networks, mapping 15255 interactions between 456 plant species and 1044 animal species in a prairie–forest transition in Western Illinois, USA (Robertson, 1929) (note that an equivalent co-extinction simulation was conducted in Memmott et al., 2004). Rather than simulating realistic extinctions, the experiments serve to compare the trajectories of different networks, and the scenarios used here (random, best and worst) are also purely hypothetical. Nonetheless, using informed scenarios based on actual species vulnerabilities provides insights into how the primary effects of global change can propagate through network links and eventually cause the loss of species not directly affected by change. For example, we might simulate future changes by removing macrophyte species based on predicted climatic conditions. As an example of the insights gained through such approaches, Strona & Bradshaw (2018) showed how secondary effects can actually be key to mass extinctions.
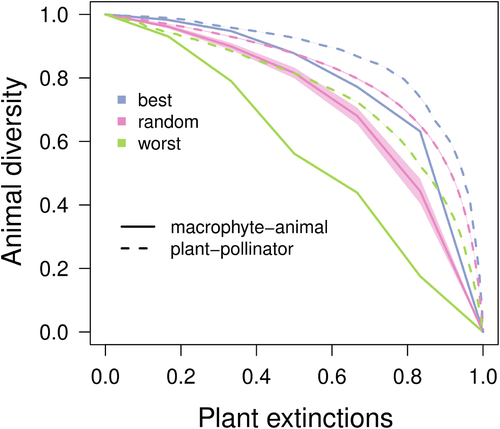
Although there are clear similarities between the co-extinction trajectories observed in the macrophyte–animal network compared to those observed in the plant–pollinator network, the former appears more fragile (Fig. 8). In other words, the macrophyte–animal network shows a faster decline in animal diversity in response to plant loss than does a plant–pollinator network. As these co-exctinction simulations are based on only two networks and on hypothetical rather than informed scenarios of species loss, the results presented cannot be generalised. However, the consistency in the observed trajectories is encouraging, and urges the exploration of the structure and robustness of macrophyte–animal networks through approaches more commonly used in terrestrial plant–animal networks. Furthermore, the results suggest that interesting differences might exist between the systems. Thus, further investigation could yield insights into the mechanisms possibly controlling differential responses to global change-induced diversity loss among aquatic and terrestrial systems (Antão et al., 2020).
(b) Effects of experimental perturbation on macrophyte–animal networks
To illustrate another application of network analysis in the study of macrophyte–animal systems under disturbance, we use data generated from experimental perturbations of rocky intertidal macroalgal canopies [Ericaria amentacea (C.Agardh) Molinari & Guiry (formerly Cystoseira amentacea Bory var. stricta Montagne)]. These experiments were originally undertaken to investigate early warning indicators of regime shifts in understorey assemblages following colonisation by algal turfs (Benedetti-Cecchi et al., 2015).
The experimental design consisted of two factors: (i) a press perturbation, i.e. the annual clipping of algal canopies (“canopy”), simulating the expected effects of rising temperatures, with four levels (a control and three increasing levels of perturbation); crossed with (ii) a pulse disturbance, i.e. a one-time-removal of invertebrates and algae (“clearing”) in 10 cm × 10 cm patches within the experimental area, simulating the effect of strong storms, with two levels (uncleared and cleared). Each combination of treatment (‘canopy’ × ‘clearing’) levels was replicated five times, and the percentage cover of understorey invertebrates and algae was sampled eight times (approximately annually) in 20 cm × 20 cm quadrats over a 7-year experimental period (2007–2013) [see Benedetti-Cecchi et al., 2015 for further details].
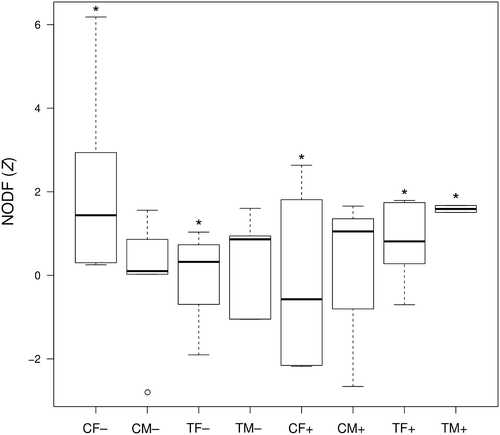
Starting from these experimental data, we generated networks of co-occurrence of understorey invertebrates and turf-forming algae, and then assessed the effect of the two disturbance factors (“canopy” and “clearing”) on network structure and properties. We first determined which species occurred in the same sampling unit more frequently than expected by chance using the package cooccur, based on the probabilistic model of species co-occurrence from Veech (2013). We hence collated all the species pairs showing significant positive co-occurrence (P < 0.05) and generated invertebrate–algae co-occurrence networks under each combination of experimental treatments and sampling dates. We then computed the nestedness of each co-occurrence network as the effect-size (Z) obtained by standardising the observed NODF value (Almeida-Neto et al., 2008) by the NODF values of 1000 null networks (using the procedure described in Section VI.2).
A linear mixed-effect model was fitted using the package nlme (Pinheiro et al., 2007) to test the correlation between network nestedness and the combination of ‘canopy’ and ‘clearing’ treatments (fixed effects), considering the sampling date as a random effect. The two most perturbed groups showed a significant correlation between NODF and treatments (‘turf margin + clearing’, t(23) = 2.42, P < 0.05; ‘turf full + clearing’, t(23) = 2.48, P < 0.05), indicating an interactive effect of canopy clipping and clearing on network nestedness (Fig. 9). The invasion of turf-forming algae in cleared and canopy-depleted plots hence appears to drive significant structural changes in the co-occurrence networks of understorey invertebrates and algae, with respect to unperturbed plots. Disturbance can have significant impacts on the architecture of ecological networks, and, in general, perturbed natural communities appear less structured than in stable environmental conditions (Vanbergen et al., 2014; de Assis Bomfim et al., 2018). Our results reflect those reported by Benedetti-Cecchi et al. (2015), which showed dramatic shifts in understorey assemblages when press perturbation and pulse disturbance are combined.
Hence, on the one hand co-occurrence networks can provide an alternative framework to assess the effect of disturbance on macrophyte–animal assemblages in experimental studies, providing complementary, ecologically relevant information to that obtainable with other analytical approaches. On the other hand, adopting ecological networks in the context of controlled perturbation experiments allows us to establish direct causal links between shifts in species co-occurrences and network properties in response to disturbance.
VII. FUTURE DIRECTIONS
The examples outlined in the previous sections show that ecological network analysis can provide a thorough understanding of patterns and processes in macrophyte–animal systems not achievable through other approaches (Section VI.2). Most notably, network analysis permits assessing the robustness of macrophyte–animal systems to disturbance and species loss (Section VI.3). In this regard, network theory is a powerful tool to assess how the structure and organisation of macrophyte–animal systems might be affected by on-going and future processes related to anthropogenic disturbance and climate change and to make predictions on future winners and losers.
As macrophyte–animal systems are embedded in littoral food webs, a future direction (anticipated in Section III) would be integrating the analysis of bipartite macrophyte–animal networks into a broader ecosystem/food-web framework. In fact, the accurate modelling of changes in macrophyte–animal links is a premise for better predicting what might happen to higher trophic levels. Clearly, this would require the additional and demanding step of identifying the links connecting animals associated with macrophytes with the other food-web consumers (e.g. fish or predatory invertebrates). Fortunately, novel techniques such as metabarcoding (e.g. used for a precise identification of fish diets through the analysis of stomach contents) make these challenges achievable (Roslin & Majaneva, 2016).
Our work is directly motivated by the ecological, societal and economic importance of macrophyte habitats and the scientific interest that they generate (Duffy et al., 2019). Reinforced by the establishment of network analysis as a primary tool to unravel ecological complexity, we have little doubt that the study of macrophyte–animal networks will flourish in the coming years. We trust that this review will help catalyse this process by revealing the full potential of a network analysis approach, and by channelling research efforts towards important open questions in ecology.
VIII. CONCLUSIONS
- We propose bipartite ecological networks as a novel analytical framework to investigate the ecological associations between marine macrophytes (i.e. macroalgae and seagrasses) and animals, on the grounds that: (i) human impacts and global environmental change are driving rapid, profound changes in macrophyte communities worldwide, with far-reaching and currently unpredictable consequences on coastal biodiversity and ecosystem functioning. (ii) In a food-web framework, basal macrophyte–animal links are generally represented with low resolution and based only on trophic interactions. Conversely, bipartite macrophyte–animal networks provide higher detail and account for the suite of interactions through which macrophytes sustain animal diversity, increasing our ability to model processes at the macrophyte–animal interface and their potential effects on coastal ecosystems as a whole.
- Through examples based on literature data, we provide guidance on how to assemble bipartite macrophyte–animal networks, explore their structure and model the effects of disturbance on network properties. We also discuss the range of ecological information obtainable with this approach.
- We suggest potential future directions, built on representing the multiple types of ecological interactions at play in macrophyte–animal systems in a multi-layer framework, and on integrating macrophyte–animal networks into coastal food-webs.




