NAB2::STAT6 fusions and genome-wide DNA methylation profiling: Predictors of patient outcomes in meningeal solitary fibrous tumors
Kathryn L. Eschbacher and Quynh T. Tran are co-first authors. Caterina Giannini and Florian Haller are co-senior authors.
Abstract
Meningeal solitary fibrous tumors (SFT) are rare and have a high frequency of local recurrence and distant metastasis. In a cohort of 126 patients (57 female, 69 male; mean age at surgery 53.0 years) with pathologically confirmed meningeal SFTs with extended clinical follow-up (median 9.9 years; range 15 days–43 years), we performed extensive molecular characterization including genome-wide DNA methylation profiling (n = 80) and targeted TERT promoter mutation testing (n = 98). Associations were examined with NAB2::STAT6 fusion status (n = 101 cases; 51 = ex5-7::ex16-17, 26 = ex4::ex2-3; 12 = ex2-3::exANY/other and 12 = no fusion) and placed in the context of 2021 Central Nervous System (CNS) WHO grade. NAB2::STAT6 fusion breakpoints (fusion type) were significantly associated with metastasis-free survival (MFS) (p = 0.03) and, on multivariate analysis, disease-specific survival (DSS) when adjusting for CNS WHO grade (p = 0.03). DNA methylation profiling revealed three distinct clusters: Cluster 1 (n = 38), Cluster 2 (n = 22), and Cluster 3 (n = 20). Methylation clusters were significantly associated with fusion type (p < 0.001), with Cluster 2 harboring ex4::ex2-3 fusion in 16 (of 20; 80.0%), nearly all TERT promoter mutations (7 of 8; 87.5%), and predominantly an “SFT” histologic phenotype (15 of 22; 68.2%). Clusters 1 and 3 were less distinct, both dominated by tumors having ex5-7::ex16-17 fusion (respectively, 25 of 33; 75.8%, and 12 of 18; 66.7%) and with variable histological phenotypes. Methylation clusters were significantly associated with MFS (p = 0.027), but not overall survival (OS). In summary, NAB2::STAT6 fusion type was significantly associated with MFS and DSS, suggesting that tumors with an ex5::ex16-17 fusion may have inferior patient outcomes. Methylation clusters were significantly associated with fusion type, TERT promoter mutation status, histologic phenotype, and MFS.
1 INTRODUCTION
Meningeal solitary fibrous tumor (SFT) is a rare tumor, typically occurring in adults, most frequently intracranially, associated with high rates of local recurrence and distant metastases even late in the disease course [1, 2]. The term SFT was ultimately adopted in the 2021 WHO Classification of Central Nervous System Tumors (CNS WHO), after a transition phase in the 2016 WHO classification recommending the use of the combined term SFT/hemangiopericytoma (HPC), and now encompasses the full spectrum of tumors previously classified as meningeal SFT and HPC [3].
Meningeal SFTs, similar to SFT across all other anatomic sites, are molecularly characterized by an inversion in chromosome 12, resulting in a NAB2::STAT6 fusion. Notably, these tumors are defined molecularly despite showing prominent histological variation, from having an SFT appearance to an HPC appearance. Fusion breakpoints in the STAT6 gene commonly occur either inside the SH2 domain or close to its N-terminus. In the NAB2 gene, they often appear after the nuclear (N) localization signal. The resulting NAB2::STAT6 fusion protein translocates to the nucleus and can, therefore, be consistently detected using STAT6 immunohistochemistry [4-8].
A variety of breakpoints have been identified within the NAB2 and STAT6 genes, NAB2 exon4::STAT6 exon2-3 (ex4::ex2-3) and NAB2 exon5-7::STAT6 exon16-17 (ex5-7::ex16-17) being the most common [9-11]. Amongst meningeal SFTs, the NAB2::STAT6 fusion breakpoints (fusion type) correlate with histologic phenotype (e.g., classic SFT vs. HPC-like), with those with ex4::ex2-3 more likely to have a classic SFT appearance and those with ex5-7::ex16-17 more likely to have an intermediate or HPC-like phenotype [1, 5, 12]. An association of the ex5-7::ex16-17 fusion type with higher-grade histologic features has also been identified [5, 13]. Up to now, however, a definite statistically significant association between fusion type and patient outcomes has not been found [1, 13, 14]. Similar findings have been reported in extracranial SFTs, including those in the soft tissue [9-11, 15, 16].
TERT promoter mutations have been found in approximately one third of meningeal SFT, but no clear association with TERT promoter mutation and patient outcomes has been identified [13]. In contrast, TERT promoter mutation has been associated with adverse outcomes or dedifferentiation in extracranial sites [16-19].
The CNS WHO grading criteria was modified in 2021 following the results of two separate studies of meningeal SFT, incorporating a strict definition based on either STAT6 nuclear expression or confirmed presence of NAB2::STAT6 fusion [1-3]. The first study by Fritchie et al. examined a cohort of 133 patients and found that necrosis was associated with recurrence-free survival (RFS) and, while NAB2::STAT6 fusion type was associated with histologic phenotype, there was no significant association with RFS or overall survival (OS) [1]. The second study by Macagno et al. evaluated a cohort of 132 patients and found that mitotic activity (≥5 mitoses/10 high-power fields) was associated with progression-free survival (PFS) and disease-specific survival (DSS), whereas necrosis was associated with DSS [2]. NAB2::STAT6 fusion type was not analyzed [2].
At present, meningeal SFT are graded, based upon histologic findings, as CNS WHO Grade 1, 2, and 3. CNS WHO Grade 1 tumors have low mitotic activity (<5 mitoses per 10 adjacent high-power fields [HPF, ×400; 1 HPF = 0.22 mm2]); CNS WHO Grade 2 are non-necrotic tumors with 5 or more mitoses per 10 HPF; and CNS WHO Grade 3 tumors additionally have necrosis [3].
In this study, we have expanded the molecular characterization of our original SFT cohort [1], performing genome-wide DNA methylation profiling and TERT promoter mutation analysis, and correlated all data, including NAB2::STAT6 fusion type, with extended patient follow-up, and in the context of 2021 CNS WHO grading.
2 MATERIALS AND METHODS
2.1 Case identification
A cohort of 126 patients with meningeal SFTs was identified from five tertiary care centers [1]. As previously described [1], tumor diagnosis was pathologically confirmed through a review of the primary tumor resection (n = 90), local recurrence (n = 35), or metastasis (n = 1). Cases were selected that demonstrated nuclear positivity for STAT6, NAB2::STAT6 fusion by sequencing, or both. Patient data regarding treatment and follow-up data (including date of original diagnosis for recurrent or metastatic patients) were collected from the medical records of the respective institutions.
2.2 NAB2::STAT6 fusion assessment
NAB2::STAT6 fusion status was successfully assessed in n = 101 cases, as previously described [1]. In brief, RNA extraction was performed using the Qiagen miRNeasy FFPE kit by methods previously described by Wang et al. [20]. Screening of the most common NAB2::STAT6 fusions was performed, including exon 4::exon 2, exon 4::exon 3, exon 6::exon 16, and exon 6::exon 17 using single-plex PCR with subsequent agarose gel electrophoresis, identifying NAB2::STAT6 gene fusions. Samples with no results by single-plex PCRs were analyzed by next-generation sequencing using the Archer FusionPlex Sarcoma Kit (Archer Dx Inc, Boulder, CO) to detect fusions of 26 genes employing the Anchored Multiplex PCR-based enrichment.
2.3 TERT promoter mutation status
Next-generation sequencing mutation analysis with a 160 gene panel was attempted (n = 100 of 109), but the majority of specimens failed quality control, likely due to paraffin block age, with evaluable results in only 26 cases (results provided in Table S1). Targeted TERT promoter mutation status was then performed and was successful in 98 (of 109) cases, utilizing a SNaPshot assay using the ABI Prism 2500 Genetic Analyzer and Snapshot-Multiplex-Kit (Applied Biosystems, Foster City, CA), as previously described, to identify hotspot mutations at positions −146 and −124 base pairs of the TERT promoter [21, 22].
2.4 DNA extraction and genome-wide DNA methylation profiling
DNA was isolated by using a QIAamp DNA FFPE Kit (Cat. No. 56404) from Qiagen. FFPE DNA quality was then analyzed by the Infinium FFPE QC Kit from Illumina (Cat. No. WG-321-1001). The DNA samples were further processed with ΔCt <5 in comparison with the quality control DNA reference from the kit. DNA concentration was quantified fluorimetrically by using Qubit. A total of 500 ng DNA was used for the bisulfite conversion by EZ DNA Methylation Kit from Zymo Research (Cat. No. D5001). The resulting bisulfite-converted DNA samples were next restored by using Infinium HD FFPE DNA Restore Kit (Cat. No. WG-321-1002). Whole genome DNA methylation screening was successfully performed in n = 80 (of 109) cases utilizing the Infinium MethylationEPIC v1.0 Kit was used (Cat. No. WG-317-1001).
Methylation data from Illumina Infinium HumanMethylationEPIC BeadChip (850K) were preprocessed using the minfi package (v.1.28.3) [23] in R, version 3.5.1 [24]. Functional normalization [25] with NOOB background correction and dye-bias normalization [26] was performed. The preprocessing step also included the calculation of β values and the detection p values. All samples had detection p values less than 0.005. In addition, probes locating to sex chromosomes, containing nucleotide polymorphism (dbSNP132 Common) within five base pairs of and including the targeted CpG-site or mapping to multiple sites on hg19 (allowing for one mismatch), as well as cross-reactive probes were removed from analysis. After filtering, there were 408,864 methylation loci remain.
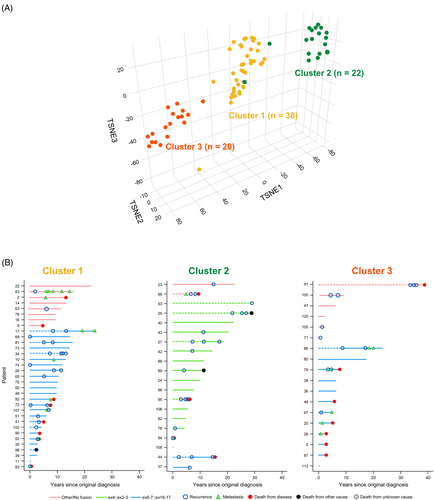
To determine the subgroups for the 80 samples, we performed an unsupervised analysis using the cola R package (v.2.2.0) [27]. We utilized the spherical kmeans (skmeans) using standard deviation as the variance metric. The k value was chosen based upon the suggested rules of: Jaccard index <0.95, if 1-PAC ≥0.90 take the maximum k; max 1-PAC, max mean silhouette, and max concordance; k = 3 gave the highest values in 1-PAC, mean silhouette, and Jaccard index (Figure S1A,B). We then performed dimensionality reduction and visualization using t-distribution stochastic neighboring embedding (t-SNE) analysis on the 80 methylation profiles to validate the three subgroups. The principal component analysis was performed on the top 20,000 most variable methylated probes. The computed principal components were then used for t-SNE analysis with these parameter settings: dims = 3, perplexity = 10, and theta = 0.8.
2.5 Gene set enrichment analysis/copy number analysis
Log FC were computed for the top 20,000 most variable methylated probes and used as the ranking metric for GSEAPreranked (v3.0) [28, 29]. Probes were mapped to gene symbols using the Illumina Annotation for EPIC arrays (ilm10b4.hg19). For probes that mapped to the same gene, only one probe with maximum (up) or minimum (down) logFC were kept for gene set enrichment analysis (GSEA). GSEA was performed to determine if the members of a given gene set were enriched among the most methylated genes for each pairwise comparison. The ranked gene lists were tested against the Hallmark and Oncogenic gene sets (v6.2).
DNA copy number was recovered from combined intensities using the conumee package [30] with reference to methylation profiles from adrenal gland tissue controls. Control samples were analyzed using the Illumina HumanMethylationEPIC BeadChip (850K). Segmentation files were combined and copy number variation (CNV) frequency plot with 0.18 threshold were generated using the conumee package [30]. Statistically significant frequent CNVs were determined using GISTIC version 2.0.23 [31]. Copy number profiles output as segments were obtained from conumee R package (as described above) and used as inputs for GISTIC2. Gain and loss were categorized with CNV values greater than 0.18 or smaller than −0.18, respectively. CNVs were also divided into those that are chromosome arm-level (defined as exceeding half of the length of a chromosome arm) and focal (shorter than this). We considered events with false discovery rate q values <0.25 as significant at 90% confidence level. An “arm-level peel-off” correction was unable to assign all CNVs in the same chromosome arm of the same sample to be part of a single event when determining whether multiple significantly recurrent events exist on that chromosome arm.
To further assess the results of the GSEA, formalin-fixed paraffin-embedded tissues in a subset of cases (n = 53) were stained with β-catenin (Cell Marque Rocklin, CA; mouse monoclonal antibody from supernatant diluted in tris-buffered saline) and LEF1 (Abcam, anti-LEF1 rabbit monoclonal antibody). β-Catenin expression was recorded as diffuse nuclear, patchy nuclear, or sparse nuclear and negative (cytoplasmic staining only). LEF1 expression was recorded as positive (50%–80% and >80%) and negative (absent, <50%).
The β values of CpG reside in two probes within a region upstream of the transcriptional start site of TERT (cg10767223 and Cg11625005) n = 80 (of 109) cases were analyzed. These probes were selected as they have been associated with TERT expression when hypermethylated [32, 33]. No normal cut-off value was available for meninges.
2.6 Classification and grading
Tumors were graded based on the most recent 2021 CNS WHO [3]. Irrespective of their histologic phenotype, tumors with fewer than 5 mitoses per 10 high-power fields (HPF, ×400; 1 HPF = 0.22 mm2) were considered CNS WHO Grade 1; tumors with 5 or more mitoses per 10 HPF without necrosis were considered CNS WHO Grade 2; tumors with 5 or more mitoses per 10 HPF and necrosis were considered CNS WHO Grade 3.
2.7 Statistical analysis
Patient demographics and tumor characteristics were summarized descriptively with frequencies, percentages, mean, median, and ranges, as appropriate. Fisher's exact tests were used to compare variables between selected groups. OS, RFS, metastasis-free survival (MFS), PFS (including recurrence or metastasis), and DSS (including death from disease) were compared between selected groups with Cox proportional hazard regression models, using the likelihood ratio test to assess significance, and 95% confidence intervals (CIs) were reported for survival estimates and hazard ratios (HRs). Depending on the variable (fusion type, methylation cluster, CNS WHO grade), statistical analysis either included the full patient cohort and was evaluated from the time of original diagnosis as available; or only the cases in which the primary tumor resection was reviewed, and in this case it was evaluated from the time of surgery (equivalent to the time of original diagnosis among primary tumors). Identification of the NAB2::STAT6 fusion type has been described as stable in metachronous recurrences from the same patient [34] and methylation-based classification has been shown to be stable across time in other tumor types [35]. Within our own cohort, two patients had identical methylation cluster results at primary resection and recurrence (one case) or between primary resection and distant metastasis (one case). Therefore, when the analysis was limited to fusion type and methylation cluster, the assumption could be made that these variables would not have changed over time. In these cases, the analysis included all patients and was considered from the time of diagnosis to limit confounding the results since the patients in whom only the non-primary resection was available had already undergone progression (recurrence or metastasis) [9]. When evaluating CNS WHO grade, the analysis included only the patients in whom primary tumor resection was reviewed and follow-up was evaluated from the time of surgery. This was necessary as, in some instances (n = 35 of 126), tissue from the primary resection was not available for grading and it could not be assumed that the grade at original diagnosis would have been the same as at recurrence [34].
When performing statistical analysis for all patients, OS was defined as the time between diagnosis and death (all causes), censoring patients still alive at last follow-up. RFS was defined as the time between diagnosis and the first local recurrence, censoring for patients without recurrence at the time of the last follow-up. MFS was defined as the time between diagnosis and the first distant metastasis, censoring for patients without distant metastasis at the time of the last follow-up. PFS was defined as the time between diagnosis and the first adverse event for the patient following surgery (local recurrence or metastasis), censoring patients without an adverse event at the time of last follow-up. DSS was defined as death specifically due to disease, while censoring deaths of unknown or other cause and patients still alive. Adjusted analyses were conducted with multivariable Cox proportional hazards regression models, limiting the number of predictors based on total available events in the data. Furthermore, molecular fusion status was dichotomized as ex5-7::ex16-17 (the most frequent) versus other fusions combined, given limited statistical power with all sparsely distributed categories. p Values less than 0.05 were considered statistically significant. All analyses were performed using SAS version 9.4 (SAS Institute Inc., Cary, NC) or R version 4.2.2 [24].
3 RESULTS
3.1 Patient demographics and outcomes
Of the 126 patients (69 male, 57 female), follow-up data was available for 124 (98%) patients; however, time of original diagnosis was unknown for one of those patients. Among those with known diagnosis date, the median follow-up time was 9.3 years (range 15 days–43 years). The mean age at time of diagnosis was 50.0 years (range 10.6–83.0 years). The mean age at time of surgery was 53.0 years (range 10.6–87.3 years). Extent of resection was known for 108 (of 126) patients with 57.4% undergoing gross total resection (n = 62) and 42.6% undergoing subtotal resection (n = 46).
At last follow-up, 84 patients were alive, including 35 with no evidence of disease, 25 with unknown disease status, and 24 with disease (progressive, stable, or metastatic). Of the deceased patients (n = 40), 29 had died from disease, 9 had died from other causes, and the cause of death was unknown for 2 patients. Follow-up status and time were unknown for 2 patients. Patient demographics are summarized in Table 1.
| All cases (N = 126) | Cases with methylation or molecular data (N = 110) | |
|---|---|---|
| Age at diagnosis (years) | ||
| Mean (SD) | 50.0 (15.8) | 50.5 (15.8) |
| Range | (10.6–83.0) | (10.6–83.0) |
| Age at surgery (years) | ||
| Mean (SD) | 53.0 (15.1) | 52.8 (15.8) |
| Range | (10.6–87.3) | (10.6–87.3) |
| Gender | ||
| Female | 57 (45.2%) | 50 (45.5%) |
| Male | 69 (54.8%) | 60 (54.5%) |
| Specimen type | ||
| Recurrent/metastasis | 36 (28.6%) | 29 (26.4%) |
| Primary | 90 (71.4%) | 81 (73.6%) |
| CNS WHO Grade | ||
| 1 | 76 (60.3%) | 61 (55.5%) |
| 2 | 36 (28.6%) | 36 (32.7%) |
| 3 | 14 (11.1%) | 13 (11.8%) |
| Necrosis | 17 (13.5%) | 15 (13.6%) |
| Histologic phenotype | ||
| HPC | 24 (19.0%) | 20 (18.2%) |
| Intermediate | 50 (39.7%) | 47 (42.7%) |
| SFT | 52 (41.3%) | 43 (39.1%) |
| TERT promoter mutation | ||
| Wild-type | 87 (88.8%) | 78 (87.6%) |
| Mutant | 11 (11.2%) | 11 (12.4%) |
| Not available | 28 | 21 |
| Methylation cluster | ||
| 1 | 38 (47.5%) | 38 (47.5%) |
| 2 | 22 (27.5%) | 22 (27.5%) |
| 3 | 20 (25.0%) | 20 (25.0%) |
| Not available | 46 | 30 |
| NAB2::STAT6 fusion type | ||
| ex5-7::ex16-17 | 51 (50.5%) | 51 (50.5%) |
| ex4::ex2-3 | 26 (25.7%) | 26 (25.7%) |
| Other or no fusion detected | 24 (23.8%) | 24 (23.8%) |
| Not available or failed | 25 | 9 |
| Follow-up status available following diagnosis, N | 124 | 108 |
| Time from diagnosis to last follow-upa | ||
| N | 123 | 107 |
| Median | 9.9 years | 9.4 years |
| Range | (15 days–43 years) | (15 days–38.8 years) |
| Events following diagnosis, N | ||
| Recurrence | 63 | 54 |
| Metastasis | 25 | 22 |
| Recurrence or metastasis | 70 | 61 |
| Status at last follow-up, N | ||
| Alive, with disease | 24 | 20 |
| Alive, without disease | 35 | 29 |
| Alive, unknown disease status | 25 | 24 |
| Death due to diseaseb | 29 | 27 |
| Death, other cause | 9 | 7 |
| Death, unknown cause | 2 | 1 |
| Unknown status, no follow-up | 2 | 2 |
- a Diagnosis date unknown for one patient among those for whom status was known.
- b Includes one patient with unknown date of diagnosis.
From time of initial diagnosis amongst patients who had died, the median OS was 28.5 years, and the median RFS was 11.3 years. When considering patients with tissue from time of initial surgery (n = 90), the median OS amongst patients who had died was 14.7 years and the median RFS was 14.9 years (Figure S2A,B).
3.2 NAB2::STAT6 fusion type
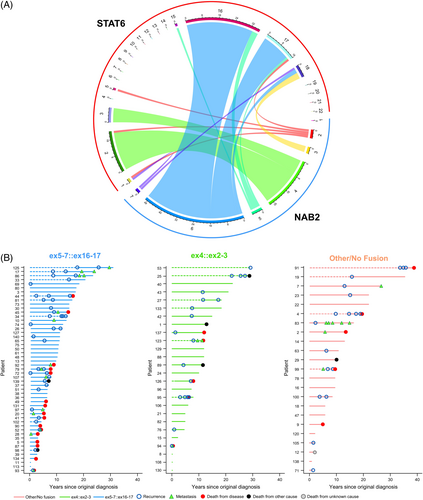
NAB2::STAT6 fusion analysis was successfully performed in 101 cases. The majority of tumors had a NAB2 exon5-7::STAT6 exon16-17 (ex5-7::ex16-17) (n = 51; 50.5%). Other fusion types detected included 26 NAB2 exon4::STAT6 exon2-3 (ex4::ex2-3; 25.7%) and 12 NAB2 exon2-3::STAT6 exonANY/other (ex2-3::exANY/other; 11.9%) (Figure 1A). No fusion was detected in 12 cases (11.9%), all of which demonstrated STAT6 nuclear expression by immunohistochemistry. A summary of patient outcomes (recurrence, metastasis, or death) across disease course categorized by fusion type can be seen in Figure 1B.
3.3 TERT promoter mutation status
Targeted TERT promoter mutation testing was successfully performed in 98 cases. TERT promoter mutations were identified in 11 cases (11.2%), including TERT c.-124G>A = 10; TERT c.-146G>A = 1. TERT promoter mutation was observed in cases with ex4::ex2-3 fusions (n = 6), ex2-3::exANY/other (n = 2), fusion negative cases (n = 2), and a case in which fusion type could not be determined (n = 1). Although 58.6% of TERT wild-type cases harbored ex5-7::ex16-17 fusions, none of the TERT promoter mutant cases had concurrent ex5-7::ex16-17 fusions (p = 0.0002) (Table 2).
| TERT promoter mutation status | |||
|---|---|---|---|
| Mutant (n = 11) | Wild-type (n = 87) | p Value | |
| NAB2::STAT6 fusion type | 0.0002 | ||
| ex5-7::ex16-17 | 0 (0%) | 41 (58.6%) | |
| ex4::ex2-3 | 6 (60.0%) | 12 (17.1%) | |
| Other or no fusion | 4 (40.0%) | 17 (24.3%) | |
| Missing/faileda | 1 | 17 | |
- Abbreviations: ex5-7::ex16-17, NAB2 exon5-7::STAT6 exon16-17; ex4::ex2-3 NAB2 exon4::STAT6 exon2-3.
- a Excluded from denominator.
3.4 Genome-wide DNA methylation profiling
Genome-wide DNA methylation profiling was successfully performed in 80 cases, revealing three distinct methylation clusters: Cluster 1 (n = 38), Cluster 2 (n = 22), and Cluster 3 (n = 20) (Figure 2A). Methylation clusters were significantly associated with fusion type (p < 0.001), with the majority of tumors in Cluster 2 (n = 16; 80.0%) having an ex4::ex2-3 fusion. Neither Cluster 1 nor Cluster 3 included cases with an ex4::ex2-3 fusion. They were composed predominantly of tumors with the ex5-7::ex16-17 fusion (Cluster 1: n = 25, 75.8%; Cluster 3: n = 12, 66.7%) (Table 3). Swimmer's plots depicting the distribution of NAB2::STAT6 fusion type amongst the methylation classes and adverse patient events across the length of follow-up are shown in Figure 2B.
| Methylation cluster | |||||
|---|---|---|---|---|---|
| 1 (n = 38) | 2 (n = 22) | 3 (n = 20) | Total (n = 80) | p Value | |
| NAB2::STAT6 fusion type | <0.0001 | ||||
| ex5-7::ex16-17 | 25 (75.8%) | 2 (10.0%) | 12 (66.7%) | 39 (54.9%) | |
| ex4::ex2-3 | 0 (0.0%) | 16 (80.0%) | 0 (0.0%) | 16 (22.5%) | |
| Other or no fusion | 8 (24.2%) | 2 (10.0%) | 6 (33.3%) | 16 (22.5%) | |
| Missing/faileda | 5 | 2 | 2 | 9 | |
| TERT promoter mutation status | 0.0002 | ||||
| TERT mutant | 0 (0.0%) | 7 (33.3%) | 1 (5.6%) | 8 (10.7%) | |
| TERT wild-type | 36 (100.0%) | 14 (66.7%) | 17 (94.4%) | 67 (89.3%) | |
| Missing/faileda | 2 | 1 | 2 | 5 | |
| 2021 CNS WHO Grade | 0.56 | ||||
| Grade 1 | 19 (50.0%) | 15 (68.2%) | 10 (50.0%) | 44 (55.0%) | |
| Grade 2 | 14 (36.8%) | 4 (18.2%) | 6 (30.0%) | 24 (30.0%) | |
| Grade 3 | 5 (13.2%) | 3 (13.6%) | 4 (20.0%) | 12 (15%) | |
- Abbreviations: CNS, central nervous system; ex5-7::ex16-17, NAB2 exon5-7::STAT6 exon16-17; ex4::ex2-3, NAB2 exon4::STAT6 exon2-3; WHO, World Health Organization.
- a Excluded from denominator.
Methylation clusters were also significantly associated with histologic phenotype (p = 0.004), with Cluster 2 encompassing predominantly tumors with a SFT phenotype (n = 15, 68.2%) and Clusters 1 and 3 being composed of a mixture of HPC-like (Cluster 1: n = 12, 31.6%; Cluster 2: n = 4, 20.0%) or intermediate (Cluster 1: n = 19, 50.0%; Cluster 2: n = 7, 35.0%) histologic phenotype (Table S2). Methylation clusters were significantly associated with TERT promoter mutation status (p < 0.001), with the majority of TERT promoter mutant cases with methylation data (n = 8) occurring in Cluster 2 (n = 7; 87.5%) (Table 3).
3.5 TERT methylation analysis
When analyzing methylation of the CpG residue at probe Cg11625005, the overall β value was significantly associated with TERT promoter mutation status (p = 0.01; median β value TERT promoter mutant = 0.08; median β value TERT wild-type = 0.14). However, there was no significant association with TERT promoter mutation status when analyzing the methylation of the CpG residue at probe cg10767223 (p = 0.55).
3.6 Gene set enrichment analysis
GSEA demonstrated that methylation Cluster 2 was enriched for genes in the WNT signaling pathway when compared to Clusters 1 and 3. To further validate this finding, immunohistochemical analysis with β-catenin and LEF1 immunohistochemical stains was performed in 53 cases (Table S3; Cluster 1 = 22, Cluster 2 = 14, Cluster 3 = 13, unknown methylation cluster = 4). β-Catenin immunohistochemistry demonstrated diffuse nuclear staining in 2 cases, patchy nuclear staining in 2 cases, sparse nuclear staining in 18 cases, and cytoplasmic (negative) staining in 31 cases. The LEF1 stain was negative in 35 cases (absent = 12, <10% = 11, 10%–50% = 12), LEF1 staining in 50%–80% of tumor cells was seen in 9 cases, and >80% staining with LEF1 was seen in 9 cases. β-Catenin and LEF1 expression did not always correlate. There was a significant association of LEF1 staining with methylation clusters (p = 0.02), with 70.6% (n = 12 of 18) of positive cases occurring in Cluster 1, 23.5% (n = 4 of 18) in Cluster 2, and 5.9% (n = 1 of 18) in Cluster 3, but a significant association was not found with fusion type (p = 0.48). There was no significant association with β-catenin nuclear expression with methylation clusters (p = 0.37) or fusion type (p = 0.48).
3.7 Copy number analysis
Aggregate CNV plots and GISTIC analysis (Figure S3) revealed that methylation Cluster 1 was enriched for focal chromosomal gains across 1p, 2, 3, 4q, 5q, 7p, 8q, 9q, 10, 11q, 12, 13q, 14q, 15q, 16q, 17, and 20 and focal losses across 1q, 4q, 6p, 10q, 11, 12, 14q, 15q, 16q, 18q, and 19q. Methylation Cluster 2 was enriched for focal chromosomal gains across 2q, 5q, 6p, 10p, 12q, 13q, 14q, 15q, and 17q and focal losses across 1q, 4q, 12p, and 16q. Methylation Cluster 3 was enriched for focal chromosomal gains across 1p, 2, 3, 4q, 5q, 7p, 8q, 9q, 10, 11q, 12, 13q, 14q, 15q, 16q, 17, and 20 and focal losses across 1q, 4q, 6p, 10q, 11, 12, 14q, 15q, 16q, 18q, and 19q.
3.8 CNS WHO grade
The cohort included 76 CNS WHO Grade 1 (60.3%), 36 Grade 2 (28.6%), and 14 Grade 3 (11.1%) tumors, according to the 2021 WHO CNS tumor classification. A comparison between the 2016 WHO and the 2021 WHO grade is shown in Sankey and Swimmer plots (Figure S4). Thirty-seven (of 37) tumors considered WHO Grade 2 in the 2016 WHO classification were “downgraded” to CNS WHO Grade 1, and 36 (of 50) tumors considered WHO Grade 3 were “downgraded” to CNS WHO Grade 2. The remaining 53 tumors had the same grade for both grading criteria (39 patients Grade 1; 14 patients Grade 3). No tumor was classified with a higher grade designation compared to the 2016 WHO grading criteria.
2021 CNS WHO grade will subsequently be referred to as CNS WHO grade. Fusion type was significantly associated with CNS WHO grade, with higher grades having relatively more patients harboring ex5-7::ex16-17 fusions (p = 0.01). Of the CNS WHO Grade 3 tumors (n = 14), 72.7% had an ex5-7::ex16-17 fusion, 18.2% had an ex4::ex2-3 fusion, and 9.1% had an ex2-3::exANY/other fusion. Amongst CNS WHO Grade 2 tumors (n = 36), 58.8% had ex5-7::ex16-17 fusions, 8.8% ex4::ex2-3, 8.8% ex2-3::exANY/other, and 23.5% the fusion type was not detected. Of the CNS WHO Grade 1 tumors, 41.1% had ex5-7::ex16-17 fusions, 37.5% exon 4::ex2-3, 14.3% ex2-3::exANY/other, and 7.1% fusion type was not detected.
There was no significant association of CNS WHO grade with TERT promoter mutation status (p = 0.23) or methylation cluster (p = 0.56) (Table 3).
3.9 Patient outcomes
3.9.1 NAB2::STAT6 fusion type
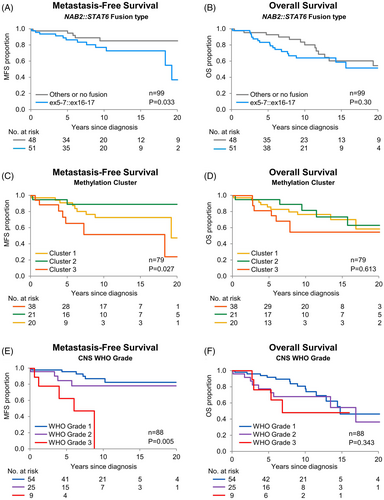
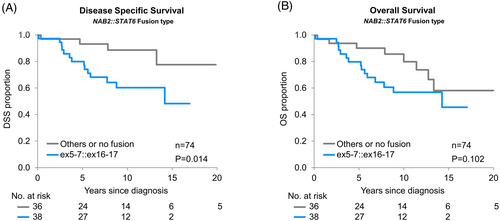
Univariate analysis of all patients from time of diagnosis revealed that, when the fusion type was dichotomized to compare cases with ex5-7::ex16-17 to all other fusion types, there was a significant association of fusion type with MFS (p = 0.03, 10-year estimates: 77.1% and 85.3% for ex5-7::ex16-17 vs. others, respectively; HR = 2.83 [95% CI: 1.09–8.28]) (Figure 3A), but not OS (p = 0.30) (Figure 3B), RFS (p = 0.15), PFS (p = 0.067), or DSS (p = 0.11). Limiting analysis to cases in which tissue from the primary resection was available (n = 74) when comparing ex5-7::ex16-17 to other fusions, there was a significant association with DSS (p = 0.01), but not OS (p = 0.10) (Figure 4A,B), MFS (p = 0.07), RFS (p = 0.69), or PFS (p = 0.35).
3.9.2 Methylation clusters
Univariate analysis of all patients from time of diagnosis demonstrated that methylation clusters were significantly associated with MFS (p = 0.03) (Figure 3C). Cluster 3 appeared to have inferior MFS compared to Clusters 1 and 2, with 10-year survival estimates of 73.7%, 89.4%, and 53.3% for Clusters 1, 2, and 3, respectively (Table S4). There was no significant association of methylation clusters with OS (p = 0.61) (Figure 3D), RFS (p = 0.91), PFS (p = 0.71), or DSS (p = 0.29).
3.9.3 TERT promoter mutation status
Univariate analysis of patients with tissue available from the primary resection from the time of surgery (n = 7 mutant, n = 62 wildtype) showed no significant association of TERT promoter mutation status with OS (p = 0.40), MFS (p = 0.99), RFS (p = 0.60), or PFS (p = 0.91).
3.9.4 CNS WHO grade
Univariate analysis of patients in which tissue was available from the primary resection of CNS WHO grade from time of surgery showed a significantly associated with MFS (p = 0.005) (Figure 3E), but no significant association with OS (p = 0.34) (Figure 3F), RFS (p = 0.95), PFS (p = 0.059), or DSS (p = 0.19). Among 76 WHO Grade 1 patients, 25 experienced recurrence, 11 metastasis, and 28 had either. Of the 36 WHO Grade 2 patients, 14 developed recurrence, 7 metastasis, and 17 either. When considering 14 WHO Grade 3 patients, 6 had recurrence, 6 metastasis, and 11 either metastasis or recurrence. The 5-year MFS was 95.9%, 84.7%, and 62.2% for WHO Grades 1, 2, and 3, respectively. Compared to WHO grade 1, the HR for metastasis was 1.87 (95% CI: 0.48–6.57) and 9.49 (95% CI: 2.63–33.18) for Grades 2 and 3, respectively.
3.10 Multivariate analysis
In multivariable Cox proportional hazards regression models that included both NAB2::STAT6 fusion type and methylation cluster (n = 71 patients with data available), fusion type was significantly associated with RFS (p = 0.04) and PFS (p = 0.03) (Table 4), but not OS (p = 0.07), DSS (p = 0.10), or MFS (p = 0.16) (Table S5). When adjusted for methylation cluster, the risk of recurrence was higher for those harboring ex5-7::ex16-17 (HR = 2.51, 95% CI: 1.03–6.59) as compared to other fusions or no fusion, with similar findings for progression (HR = 2.45, 95% CI: 1.08–6.01).
| Event-free rates, % (95% CI) | Univariate | Adjusteda | |||||
|---|---|---|---|---|---|---|---|
| Event type (N events)b | Predictor | 5-year | 10-year | HR (95% CI) | p Value | HR (95% CI) | p Value |
| Recurrence (37) | Cluster 1 | 68.1 (51.7, 84.5) | 41.3 (22.8, 59.7) | Reference | Reference | ||
| Cluster 2 | 73.7 (53.9, 93.5) | 55.3 (31.9, 78.6) | 0.96 (0.45, 2.00) | 0.920 | 1.70 (0.66, 4.51) | 0.276 | |
| Cluster 3 | 65.9 (40.8, 90.9) | 43.9 (5.0, 82.8) | 0.75 (0.27, 1.85) | 0.553 | 0.77 (0.27, 1.88) | 0.580 | |
| No fusion/other fusion | 73.8 (58.2, 89.4) | 61.5 (43.3, 79.7) | Reference | Reference | |||
| ex5-7::ex16-17 | 65.5 (49.5, 81.6) | 30.8 (12.3, 49.3) | 1.74 (0.87, 3.57) | 0.116 | 2.51 (1.03, 6.59) | 0.042 | |
| Recurrence or metastasis (42) | Cluster 1 | 68.1 (51.7, 84.5) | 30.3 (13.0, 47.5) | Reference | Reference | ||
| Cluster 2 | 68.0 (46.9, 89.2) | 55.6 (32.4, 78.9) | 0.80 (0.38, 1.61) | 0.536 | 1.39 (0.56, 3.49) | 0.478 | |
| Cluster 3 | 54.9 (29.4, 80.4) | 36.6 (2.7, 70.5) | 0.88 (0.36, 1.95) | 0.754 | 0.90 (0.37, 2.00) | 0.802 | |
| No fusion/other fusion | 70.1 (53.7, 86.5) | 57.9 (39.5, 76.4) | Reference | Reference | |||
| ex5-7::ex16-17 | 60.5 (44.1, 76.8) | 20.7 (5.0, 36.3) | 1.98 (1.03, 3.93) | 0.039 | 2.45 (1.08, 6.01) | 0.032 | |
- Abbreviation: ex5-7::ex16-17, NAB2 exon5-7::STAT6 exon16-17.
- a From models that include both methylation cluster and dichotomized fusion type together.
- b Considering events from time of original diagnosis to last follow-up.
Because CNS WHO Grade 3 was associated with higher likelihood to harbor ex5-7::ex16-17, multivariate Cox proportional hazards regression models was utilized to include fusion along with CNS WHO grade. When adjusting for fusion type in patients in which tissue from the primary resection was available (n = 74), MFS and PFS were significantly worse for WHO Grade 3 versus 1 (p = 0.01 [HR = 6.97] and 0.02 [HR = 4.46], respectively; Table 5). There was no significant association between WHO grade with OS (p = 0.28), DSS (p = 0.36), or RFS (p = 0.95) (Table S6).
| Event-free rates, % (95% CI) | Univariate | Adjusteda | |||||
|---|---|---|---|---|---|---|---|
| Event type (N events)b | Predictor | 5-year | 10-year | HR (95% CI) | p Value | HR (95% CI) | p Value |
| Death from disease (18) | No fusion/other fusion | 93.3 (84.3, 100.0) | 88.7 (76.3, 100.0) | Reference | Reference | ||
| ex5-7::ex16-17 vs. other | 80.0 (66.7, 93.3) | 60.1 (42.8, 77.4) | 3.57 (1.28, 12.63) | 0.014 | 3.16 (1.10, 11.34) | 0.031 | |
| CNS WHO Grade 1 | 92.3 (84.0, 100.0) | 78.6 (64.1, 93.0) | Reference | Reference | |||
| CNS WHO Grade 2 | 79.0 (60.6, 97.5) | 67.8 (46.3, 89.2) | 2.08 (0.73, 5.79) | 0.166 | 1.71 (0.59, 4.82) | 0.313 | |
| CNS WHO Grade 3 | 68.6 (32.1, 100.0) | 51.4 (11.5, 91.4) | 3.06 (0.67, 10.62) | 0.135 | 2.45 (0.53, 8.70) | 0.226 | |
| Metastasis (13) | No fusion/other fusion | 97.1 (91.4, 100.0) | 88.4 (75.9, 100.0) | Reference | Reference | ||
| ex5-7::ex16-17 vs. other | 85.1 (73.0, 97.3) | 70.6 (52.4, 88.7) | 3.10 (0.92, 13.96) | 0.068 | 2.23 (0.61, 10.54) | 0.231 | |
| CNS WHO Grade 1 | 94.9 (88.0, 100.0) | 87.6 (75.9, 99.2) | Reference | Reference | |||
| CNS WHO Grade 2 | 89.3 (74.9, 100.0) | 81.8 (62.6, 100.0) | 1.46 (0.30, 5.97) | 0.610 | 1.25 (0.25, 5.20) | 0.770 | |
| CNS WHO Grade 3 | 71.4 (38.0, 100.0) | 0.0 (Non-estc) | 9.24 (2.20, 37.01) | 0.004 | 6.97 (1.58, 29.69) | 0.012 | |
- Abbreviation: ex5-7::ex16-17, NAB2 exon5-7::STAT6 exon16-17.
- a From models that include both WHO and dichotomized fusion together.
- b Considering events from time of original diagnosis to last follow-up.
- c Confidence interval cannot be estimated due to lack of variability.
Considering fusion type in these same models, after adjusting for CNS WHO grade, those with ex5-7::ex16-17 had significantly worse DSS as compared to the others (p = 0.03 [HR = 3.16], Table 5). There was no significant association of fusion type with OS (p = 0.18), RFS (p = 0.76), MFS (p = 0.23), or PFS (p = 0.75) after adjusting for CNS WHO grade (Table S5). Figure S5 depicts Swimmers plots including patient's in which tissue from the primary resection and molecular data was available (n = 75) depicting adverse patient events comparing ex5-7::ex16-17 to other fusion types (A) and comparing ex5-7::ex16-17 to other fusion types by 2021 WHO grade (B).
4 DISCUSSION
The rarity of meningeal SFT, a tumor with a frequency well below 1% among primary CNS tumors [36], makes it challenging to gather patient cohorts with detailed clinical information, long-term patient follow-up, and extensive molecular characterization. Our cohort (n = 126), which includes patients from five tertiary centers, provides extended follow-up and in-depth molecular analysis in a well-characterized patient cohort.
Although all SFTs are characterized by the presence of a NAB2::STAT6 fusion, our study raises the consideration of the impact of the fusion breakpoints (fusion type) on the biologic behavior of these tumors and patient outcomes. In our cohort, the fusion type (ex5-7::ex16-17 vs others) was significantly associated with MFS and with DSS when adjusting for CNS WHO grade upon multivariate analysis. These findings are similar to what was found in prior studies of SFTs in extracranial locations [9, 11, 16] and also in meningeal SFTs in which only trends were identified toward more aggressive behavior in tumors lacking the ex4::ex2-3 fusion [5, 13, 14]. In addition, fusion type was associated with CNS WHO grade (p = 0.01), with the majority of CNS WHO Grade 3 tumors (72.7%) having an ex5-7::ex16-17 fusion, while Grades 1 and 2 had fewer patients with this fusion type (58.8% and 41.1%, respectively).
The incidence of TERT promoter mutations in our cohort (11.2%, all patients) was lower than previously reported by Vogels et al. who identified TERT promoter mutations in approximately one third of their meningeal SFT cohort [13]. Similar to the study by Vogels et al., we did not find a significant association of TERT promoter mutation with patient outcome [13], which differs from findings in other anatomic locations [16-19]. However, in contrast to that study, we did find a significant association between TERT promoter mutation and fusion type (p = 0.0002), with none of the TERT promoter mutant cases harboring concurrent ex5-7::ex16-17 fusions. Given the association between TERT promoter mutation and NAB2::STAT6 fusion type and the lower proportion of TERT promoter mutant cases, these two cohorts, despite all being meningeal SFTs, likely have a different distribution in terms of NAB2::STAT6 fusion type and DNA methylation profiles. This raises the importance of having detailed molecular information available when interpreting the data from these cohorts and putting it into clinical context.
Within our cohort, we identified three separate methylation clusters amongst meningeal SFTs, which were associated with fusion type, TERT promoter mutation status, histologic phenotype, and MFS. Cluster 2 was the most distinct, with the majority of tumors (80%) having an ex4::ex2-3 fusion and an SFT histologic phenotype (68.2%). In addition, of our TERT promoter mutant cases with DNA methylation data (n = 8), 87.5% (n = 7) were within Cluster 2. Clusters 1 and 3 were less distinct, with both clusters predominantly composed of tumors with ex5-7::ex16-17 fusions (75.8% and 66.7%, respectively) and neither having tumors with ex4::ex2-3 fusions. Despite the molecular and histologic overlap between methylation Clusters 1 and 3, Cluster 3 appeared to have inferior MFS. GSEA demonstrated that Cluster 2 was enriched for genes within the WNT signaling pathway; however, we were unable to validate this finding utilizing immunohistochemical stains for β-catenin and LEF1. Although there was a significant association between positive LEF1 immunohistochemical staining and methylation cluster, this was due to the number of positive cases in Cluster 1 rather than Cluster 2. These findings highlight that there are further molecular drivers in meningeal SFT that remain to be determined.
In the present CNS tumor classification, meningeal SFT are placed in three grades based upon histologic findings. Despite the adjustments made and the improvements in the diagnosis, the predictive power of the CNS WHO scheme remains suboptimal. The long-term follow-up of our patient cohort confirms how, independently of CNS WHO grade, meningeal SFT has a high frequency of recurrence and even patients with CNS WHO Grade 1 and Grade 2 tumors may die of disease early or late in their clinical course, casting doubts if meningeal SFT should ever be considered a CNS WHO Grade 1 tumor. At the time the 2021 WHO guidelines were prepared, caution in excluding “CNS WHO Grade 1 SFT” was motivated by the largely “tertiary referral center” nature of the patient cohorts, and the fact that patients were often referred at time of recurrence. The possibility that there could be a subset of patients with CNS WHO Grade 1 meningeal SFT with a favorable outcome could not be completely excluded. In the present series, CNS WHO grade is not significantly associated with RFS or OS. It is instead significantly associated with MFS, and CNS WHO Grade 3 tumors, now a minority of the cases (compared to the WHO 2016 grading approach), present early with metastatic disease.
Limitations of our study included its retrospective nature and the variability of available clinical, treatment and follow-up information. In addition, given the age of the diagnostic material, especially in the patients with long-term follow-up, the study was limited by the number of tumors with full molecular characterization, including DNA methylation analysis (n = 80). We also had material from the primary resection available only in a subset of cases (n = 90), which impacted our ability to fully evaluate CNS WHO grading effect. Previous studies have demonstrated histologic progression at time of tumor recurrence, so that we could not infer CNS WHO grade of the primary tumor [34]. An additional limitation of our study is that extent of resection status was known only in a portion of our cases (n = 108) and included a mixture of gross total resections (n = 62) and subtotal resections (n = 46), and therefore grade may have been underestimated in some cases due to sampling bias.
Despite these limitations, our findings suggest that the molecular differences amongst meningeal SFTs, in particular fusion type and methylation cluster, have impacts on patient outcomes. This further prompts the consideration that NAB2::STAT6 fusion type may play a larger role in the natural history of these tumors and raises the question that perhaps a grading scheme that incorporates both histologic criteria and molecular criteria may help better stratify patients. However, further detailed clinical follow-up and studies of larger cohorts are needed to understand the interactions between the molecular drivers of meningeal SFTs and patient outcomes.
AUTHOR CONTRIBUTIONS
This study was designed and data was generated by Kathryn L. Eschbacher, Quynh T. Tran, Evgeny A. Moskalev, Robert Stoehr, Brent A. Orr, Florian Haller, and Caterina Giannini. Patient information and materials were supplied by Kathryn L. Eschbacher, Karen Fritchie, Andrew Guajardo, Sandro Santagata, Sabrina Rossi, Fabio Ferrarese, David R. Raleigh, Minh P. Nguyen, John Gross, Jose Velazquez Vega, Fausto Rodriguez, Arie Perry, Maria Martinez-Lage, and Caterina Giannini. The data was analyzed and the manuscript was written by Kathryn L. Eschbacher, Quynh T. Tran, Evgeny A. Moskalev, Sarah Jenkins, Brent A. Orr, Florian Haller, and Caterina Giannini. All authors contributed to the editing and revision of the manuscript.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Katelyn Reed who aided in the organization and transportation of histologic slides and materials for this project. Internal departmental funding was provided from the Mayo Clinic Department of Anatomic Pathology. No external funding was recieved for this project.
CONFLICT OF INTEREST STATEMENT
Unrelated to this work, Priscilla K. Brastianos has consulted for Advise Connect Inspire, Axiom Healthcare Strategies, CraniUS, Genentech, InCephalo Therapeutics, Kazia, MPM Capital Advisors, Sintetica, Voyager Therapeutics, Angiochem, Dantari, Elevatebio, Eli Llly, Merck, Pfizer, SK Life Sciences and Tesaro, and has received Speaker's Honoraria from Merck, Eli Lilly, Genentech-Roche, and Medscape and institutional research funding (to MGH) from Merck, Eli Lilly, Kinnate, and Mirati. Other authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




