Late Cenozoic diversification of the austral genus Lagenophora (Astereae, Asteraceae)
Abstract
Lagenophora (Astereae, Asteraceae) has 14 species in New Zealand, Australia, Asia, southern South America, Gough Island and Tristan da Cunha. Phylogenetic relationships in Lagenophora were inferred using nuclear and plastid DNA regions. Reconstruction of spatio-temporal evolution was estimated using parsimony, Bayesian inference and likelihood methods, a Bayesian relaxed molecular clock and ancestral area and habitat reconstructions. Our results support a narrow taxonomic concept of Lagenophora including only a core group of species with one clade diversifying in New Zealand and another in South America. The split between the New Zealand and South American Lagenophora dates from 11.2 Mya [6.1–17.4 95% highest posterior density (HPD)]. The inferred ancestral habitats were openings in beech forest and subalpine tussockland. The biogeographical analyses infer a complex ancestral area for Lagenophora involving New Zealand and southern South America. Thus, the estimated divergence times and biogeographical reconstructions provide circumstantial evidence that Antarctica may have served as a corridor for migration until the expansion of the continental ice during the late Cenozoic. The extant distribution of Lagenophora reflects a complex history that could also have involved direct long-distance dispersal across southern oceans. © 2014 The Linnean Society of London, Botanical Journal of the Linnean Society, 2015, 177, 78–95.
Introduction
With more than 20 000 species and a cosmopolitan distribution, Asteraceae is probably the largest family of flowering plants. The discovery of a remarkably well-preserved fossil from Eocene deposits in Argentina suggests that the family was part of an ancient flora that inhabited southern Gondwana before the establishment of oceanic barriers to dispersal (Bremer & Gustafsson, 1997; Katinas et al., 2007; Barreda et al., 2010, 2012). By the Miocene, the family had a cosmopolitan distribution (Scott, Cadman & McMillan, 2006; Barreda et al., 2010; Zavada & Lowrey, 2010). Members of the family are presently found in virtually all biomes, with the exception of Antarctica (Funk et al., 2005). The fruits of Asteraceae are one-seeded cypselae and many species have a modified pappus structure adapted for seed dispersal over great distances.
Notably, only a few genera of Asteraceae occur in Australia, New Zealand and southern South America, occasionally extending to Asia (Allan, 1961; Moreira-Muñoz & Muñoz-Schick, 2007; Ezcurra, Baccala & Wardle, 2008). One such genus is Lagenophora Cass. (Astereae). Its austral distribution, unusual in Asteraceae, makes this genus a fascinating object of study that could provide valuable new insights into the evolutionary processes that explain extant austral distribution patterns.
The fruit morphology of Lagenophora is also unusual (see below) in Asteraceae, and could be key to inferring dispersal mechanisms and biogeographical patterns in the Southern Hemisphere. Lagenophora also has an association with southern beech forest (Nothofagaceae), an important component of austral floras and the focus of much discussion about Southern Hemisphere biogeography.
Lagenophora taxonomic history, distribution and habitats
Lagenophora previously included Australasian, Hawaiian, Central American and South American species. However, Cabrera (1966) excluded the Hawaiian species from Lagenophora and placed them in Keysseria Lauterbach. Cabrera (1966) also described the new section Lagenophora section Pseudomyriactis Cabrera to include the Central American and one Venezuelan species of Lagenophora. Cabrera with Beaman & de Hong (1965) noticed the anomalous morphological characters of the species of this section. Later, Velez (1981) and Cuatrecasas (1986) excluded the Central American species and the Venezuelan species from Lagenophora and placed them in Myriactis Less. This narrowed circumscription of Lagenophora without the Central American, Venezuelan and Hawaiian species was followed by different authors (e.g. Nesom, 2001; Brouillet et al., 2009; Sancho & Pruski, 2011) and supported by recent molecular phylogenetic analyses. Indeed, Keysseria and Myriactis proved to be distantly related to Lagenophora [Brouillet et al., 2009; Nakamura et al., 2012, with Myriactis panamensis (S.F.Blake) Cuatrec. as ‘Lagenophora panamensis S.F. Blake']. Recently, Nakamura et al. (2012), based on internal transcribed spacer (ITS) sequence data, showed a paraphyletic Lagenophora closely related to Solenogyne Cass., although the authors did not make decisions on the circumscription of Lagenophora. In addition, this study did not include the southern South American Lagenophora spp. [one of which, L. nudicaulis (Comm. ex Lam.) Dusén, is the type species of the genus], which are important to completely understand the phylogenetic relationships and spatio-temporal evolution of this genus.
As presently circumscribed, Lagenophora comprises 14 species (Table 1) (Cabrera, 1966; Drury, 1974; Cuatrecasas, 1986; Nesom, 2001) and has most of its diversity in New Zealand (nine species; de Lange & Rolfe, 2010); Australia has four species [two, L. montana and L. stipitata, shared with New Zealand]; one of the Australian species extends to Asia and three species live in southern South America, including Juan Fernández and surrounding islands, Staten Island (Isla de los Estados) and the Falkland Islands (Islas Malvinas) (Fig. 1A, B). One of the South American species, L. nudicaulis, also occurs on Gough Island and the islands of Tristan da Cunha. Seven of the nine species in New Zealand are endemic. The New Zealand species occupy habitats from coastal to near the lower limit of alpine zones up to about 1300 m, and range from the Subantarctic Island groups, such as Auckland and Campbell Islands, to the subtropical Kermadec Islands (Fig. 2A). Notably, two New Zealand endemic species (L. strangulata and L. pinnatifida) are more or less restricted to southern beech forest (Fig. 2B). Lagenophora in South America has a south-temperate distribution, mostly in Andean regions (Fig. 2C), at elevations ranging from 1500 m in the north of the range to sea level at its southernmost extreme in the islands close to Tierra del Fuego (Fig. 2D). South American Lagenophora spp. are restricted to specific environments, usually as elements of understorey and light gaps of southern beech forest (Fig. 2C).
| Lagenophora species | Distribution |
|---|---|
| L. barkeri Kirk | NZ (SI) |
| L. cuneata Kirk | NZ (NI, SI) |
| L. gracilis Steetz | South-eastern AS, AU (south-eastern AU, TA), India, Java, New Caledonia, New Guinea, Philippines, Sri Lanka, Sumatra, Timor |
| L. hariotii Franch. | AR (Chubut, Neuquén, Río Negro, Santa Cruz, Tierra del Fuego), CH (VII, VIII, IX, X, XI, XII, Juan Fernández Island) |
| L. hirsuta Poepp. ex Less. | AR (Neuquén, Río Negro), CH (VII, VIII, IX, X, XII) |
| L. huegelii Benth. | AU (TA, south-eastern AU, WAU) |
| L. lanata A.Cunn. | NZ (NI) |
| L. montana Hook.f. | AU (south-eastern AU, TA), NZ (NI, SI) |
| L. nudicaulis (Comm. ex Lam.) Dusén | AR (Neuquén, Río Negro, Tierra del Fuego), CH (VI, VII, VIII, IX, X, XI, XII), Islands: Staten (de los Estados), Falklands (Malvinas), Gough, Tristan da Cunha |
| L. petiolata Hook.f. | NZ (AUI, CAI, CHI, KI, NI, SI, STI) |
| L. pinnatifida Hook.f. | NZ (NI, SI) |
| L. pumila (Forst.f.) Cheesem. | NZ (CHI, KI, NI, SI, STI) |
| L. stipitata (Labill.) Druce | AU (south-eastern AU, TA), NZ (NI), Papua |
| L. strangulata Colenso | NZ (NI, SI) |
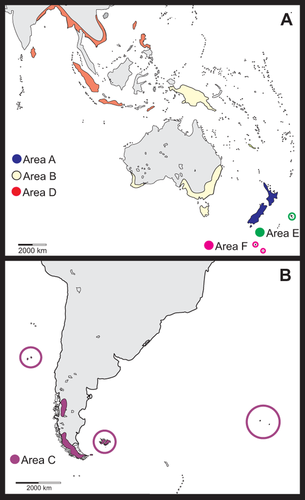
Distribution of Lagenophora. A, West Pacific distribution; B, South America, Tristan da Cunha and Gough Island. In colour, areas defined in the biogeographical analysis (when present). (A) and (B) represent Mollweide equal-area map projections.

Lagenophora habitats. A, Subalpine tussockland, New Zealand. B, Beech forest, New Zealand. C, Beech forest, South America. D, Coastal areas of Chile with scattered low beech forest. Lagenophora hariotii: E, plant habit. Lagenophora petiolata: F, epappose, beaked glandular cypsela. Lagenophora hariotii: G, ‘infructescence’ umbrella-like. (Photographs: A, by S. Wagstaff; B, by P. Fritsch; C, E–G, by G. Sancho; D, by M. Bonifacino).
Lagenophora and dispersal patterns of austral flora
Lagenophora spp. are mostly delicate plants, usually rhizomatous, that produce solitary capitula on long, often wiry scapes (Fig. 2E); the pappus, the most common dispersal structure in Asteraceae, is lacking, which is unusual (Fig. 2F). Sanmartín, Wanntorp & Winkworth (2007) suggested that dispersal mechanisms are important for inferring dispersal patterns (i.e. direct wind or water dispersal) that explain biogeographical scenarios of trans-Pacific flora. According to Sanmartín & Ronquist (2004), the dominant pattern in plants (southern South America (Australia, New Zealand)) is better explained by dispersal than by vicariance associated with the breakup of Gondwanaland and subsequent continental drift. For Asteraceae, the break-up of Gondwanaland would predate many of the existing genera.
Other authors (Ashworth et al., 2007) specifically suggested that birds might have facilitated biotic exchange and transportation among Antarctica, New Zealand, Tasmania and South America during the mid-Miocene warm interval.
The lack of a pappus in Lagenophora has intrigued researchers (Cabrera, 1966; Carlquist, 1967; Heads, 1999; Moreira-Muñoz, 2007; Moreira-Muñoz & Muñoz-Schick, 2007) and has stimulated controversial discussions about its trans-Pacific distribution. Alternative scenarios have been suggested for austral members of Asteraceae that lack a pappus. These include tectonic events (Heads, 1999, 2012) and a complex history of migration along former Antarctic coastlines and long-distance dispersal (Swenson & Bremer, 1997; Wagstaff, Breitwieser & Swenson, 2006). The debate has even considered the value of certain calibration points for inferring divergence times (Heads, 2012; Swenson, Nylinder & Wagstaff, 2012). There is an increasing need for more researchers to undertake basic data collection and interpretation to generate well-grounded hypotheses of the history of austral taxa. DNA sequence data have been used to demonstrate that trans-oceanic dispersal into and out of southern South America had an important historical role in producing the contemporary distribution of many plant and animal groups (Sanmartín & Ronquist, 2004; Wilf et al., 2012), and biome conservation appears to be the strongest filter influencing establishment, at least for plants (Crisp et al., 2009). In the light of the increasing controversy about the possible scenarios of the Southern Hemisphere trans-oceanic distributions, new information provided by molecular studies, as presented here for Lagenophora, is needed to better understand intercontinental biogeographical patterns.
Crisp, Trewick & Cook (2011) argued for a more rigorous approach to the analysis of biogeography. Rather than a biogeographical narrative, they suggested that the research questions should be framed as hypotheses. Biogeographical scenarios thus become testable. We adopted the approach that they advocated. We have undertaken a global phylogenetic analysis of Lagenophora including the South American species for the first time. We aim to use the inferred phylogenetic relationships to estimate divergence times and to test competing hypotheses of the disjunct distribution patterns in this genus. A vicariance hypothesis based on the break-up of Gondwana would be rejected if the estimated divergence times in Lagenophora were substantially more recent than the separation of Australia, southern South America and Antarctica. Alternatively, Antarctica may have served as a corridor for migration until the expansion of the continental ice shield (Miocene–Pliocene) forced lineages of Lagenophora to migrate northwards into Australia, New Zealand, South America and Asia. If gene flow was relatively unrestricted among the Antarctic source populations of Lagenophora, we predict that the early-branching lineages would be unresolved. More recent direct trans-oceanic dispersal probably also accounts for extant distribution patterns. We suggest that austral disjuncts with divergence times that are more recent than the age of the Antarctic ice shield are evidence of direct long-distance dispersal. Because the sister lineages are geographically isolated following trans-oceanic dispersal, we predict that these lineages would be phylogenetically distinct, depending on how recently the long-distance dispersal event had occurred. Finally, we predict that geological and climatic changes during the Pliocene and Pleistocene occurred in conjunction with the crown radiations in lineages of Lagenophora and may have been the important drivers of speciation.
Material and Methods
Taxon sampling
We included all 14 Lagenophora spp. (Cabrera, 1966; Drury, 1974; Cuatrecasas, 1986; Nesom, 2001; de Lange & Rolfe, 2010) in our study. Four (of four) species of Solenogyne and one (of c. 12) species of Keysseria (see Appendix 1), also belonging to subtribe Lagenophorinae, were also sequenced. Additional outgroups from tribe Astereae (Asteroideae) were selected according to Brouillet et al. (2009): species of Brachyscome Cass., Celmisia Cass., Chiliotrichum Cass., Olearia Moench, Pleurophyllum Hook.f and Vittadinia A.Rich. More distant outgroups were two representatives from other tribes of Asteroideae: Abrotanella Cass. (unassigned tribe) and Cotula L. (Anthemideae). We rooted the analysis with Dasyphyllum diacanthoides (Less.) Cabrera (Barnadesioideae). Additional available Lagenophora and Solenogyne ITS and trnK sequences (13) were obtained from GenBank (http://www.ncbi.nlm.nih.gov/GenBank/) and the corresponding outgroup sequences (see Appendix 1). The 49 newly generated sequences were deposited in GenBank and the aligned matrices in TreeBASE (Study accession = S16529).
DNA isolation, amplification and sequencing
Total DNA was extracted from leaf material (dried in silica gel or from herbarium specimens) using the DNeasy plant mini kit (Qiagen Corp., Valencia, CA, USA). We targeted the nuclear ribosomal ITS and ETS (external transcribed spacer) regions and the plastid intergenic spacer regions trnL–trnF (trnL) and the 5′trnK/matK (trnK). These regions have been proven to be useful for the reconstruction of species- and genus-level phylogenetic trees in tribe Astereae (e.g. Sancho & Karaman-Castro, 2008; Karaman-Castro & Urbatsch, 2009; Wagstaff, Breitwieser & Ito, 2011; Bonifacino & Funk, 2012; Nakamura et al., 2012). For the amplification, we used the primers given in Wagstaff et al. (2011). Polymerase chain reaction (PCR) mixes for all markers included 12 μL of sterile H2O, 1 μL of DNA, 2.5 μL of PCR buffer, 2.5 μL of deoxynucleoside triphosphates (dNTPs), 2.5 μL of each primer, 1.5 μL of MgCl2 and 0.2 or 0.4 μL of Invitrogen Taq polymerase (Life Technologies, Brazil). When necessary, dilutions of total DNA were conducted. All 25-μL PCRs were performed in a Thermal Cycler GenePro. The PCR samples were heated to 94 °C for 3 min. The double-stranded PCR products were produced via 30 cycles of denaturation (94 °C for 1 min), primer annealing (48–55 °C for 1 min), then a extension (72 ºC for 1 min) and a final extension cycle (72 °C for 2 min) followed the 30th cycle to ensure the completion of all novel strands. The PCR products were purified using a QIAquick purification kit (Qiagen Corp.). The cleaned PCR products then were labelled with fluorescent dyes (BigDye Terminator version 3.1, Applied Biosystems, Foster City, CA, USA). The reactions were sequenced in both directions by Macrogen Inc. (South Korea).
Amplification products for the four markers were obtained from the same voucher. In cases in which a marker proved difficult to amplify (e.g. L. petiolata), we tried using additional accessions (see Appendix 1). When it was not possible to amplify a given region, these bases were scored as missing data in the combined matrix. The sequence contigs were compiled and edited using Sequencher version 4.8 (Gene Codes Corporation, Ann Arbor, MI, USA). The data matrices (with additional sequences from GenBank) were aligned manually using BioEdit (Ibis Biosciences, Carlsbad, CA, USA). The plastid marker indels were not included as separate characters in the analysis.
Phylogenetic analyses
The incongruence length difference test (Farris et al., 1994) and the Shimodaira and Hasegawa test (Shimodaira & Hasegawa, 1999), as implemented in PAUP* v.4.0b10 (Swofford, 2002), were used to investigate possible incongruence between nuclear and plastid datasets. For the incongruence length difference test, we excluded uninformative characters and used the following options: heuristic search with 1000 replications of random addition sequence (RAS) + tree bisection–reconnection (TBR), saving ten trees in each replicate.
The nuclear (ITS, ETS) and plastid data (trnK and trnL regions) were analysed separately and together using maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI). Four species (L. gracilis, L. huegelii, Solenogyne bellioides Cass. and S. mikadoi Koidz.) were represented in the combined matrix only by ITS and/or trnK sequences (see Appendix 1).
The MP phylogenetic tree was reconstructed with TNT v.1.1 (Goloboff, Farris & Nixon, 2008) using the driven search option with the default settings for sectorial searches, ratchet, tree drifting and tree fusing, with ten initial random addition sequences and terminating the search after finding minimum length trees. All characters were equally weighted and unordered. Gaps were regarded as missing data. Support for clades was estimated by bootstrap analyses [bootstrap support (BS); Felsenstein, 1985] with 1000 replicates. For ML and BI analyses, jModeltest v.0.1.1 (Posada, 2008), under the Akaike information criterion (AIC), was used to determine the substitution model that best fitted the Lagenophora sequence data of each data partition and the combined dataset (see Appendix 2).
ML (Felsenstein, 1973) bootstrap analyses of the combined matrix, using Garli v.2.0 (Zwickl, 2006) under the GTR + G + I model, were executed with 100 pseudoreplicates, each based on two search replicates.
Bayesian analyses for individual data partitions were undertaken using MrBayes v.3.1.2 (Ronquist & Huelsenbeck, 2003). The searches were conducted for 2 000 000 generations with four Monte Carlo Markov chains (MCMC), three heated and one cold. The chain sample frequency and tree diagnosis were calculated every 100 generations. The average standard deviation of the split frequencies was < 0.01, and the potential scale reduction factor approached 1.0 for all parameters. The initial 25% of saved trees were discarded as burn-in and the consensus and posterior probabilities (PP) were calculated based on the remaining trees.
Bayesian analysis for the combined dataset was undertaken using Beast v.1.7.5 (Drummond et al., 2012). The assumption of a strict molecular clock was rejected by a likelihood ratio test (LRT; Felsenstein, 1988), and so divergence times were calculated with a relaxed, uncorrelated, log-normal molecular clock model using Beast v.1.7.5. Two independent MCMC searches were undertaken. The BI and dating runs in Beast v.1.7.5 of the concatenated dataset used the same matrix as employed in MrBayes analysis, with nuclear ETS and ITS and plastid trnK and trnL sequences for 31 taxa. The model set in the analysis was GTR + I + G and four gamma categories. Analyses in Beast v.1.7.5 used a speciation model that followed a Yule tree prior. MCMC chains were run for 50 million generations (burn-in 10%), with parameters sampled every 5000 generations. Tracer v.1.5 (Rambaut & Drummond, 2009) was used to assess the relevant estimated parameters and node ages (the effective sample sizes for all parameters were above 200).
Dating analyses
The potential sources of error in molecular dating analyses were pointed out by Ho & Phillips (2009). The selection and assignment of calibration points for age estimation are critical (e.g. Forest, 2009; Heads, 2012; Sauquet et al., 2012; Swenson et al., 2012), as is the choice of substitution model. In spite of the controversy about dating methods, age estimation from molecular sequences has emerged as a widely used tool for inferring when a plant lineage arrived in a particular area (Renner, 2005). In this respect, most authors agree that, even if reliable age constraints are available and incorporated in the analysis, large uncertainties remain. The use of multiple calibrations may help to accommodate among-lineage rate variation (e.g. Ho & Phillips, 2009; Sauquet et al., 2012).
To estimate divergence times, we assigned two calibration points which represent age constraints at the basal grade of the family (a fossil) and at a node near the tips of the tree (geological calibration). For the first calibration point, close to the root of the tree, we assigned a recently discovered fossil allied to Mutisioideae, which was dated to the middle Eocene in southern Argentina (Barreda et al., 2010), 47.5 Mya. According to Sauquet et al. (2012), macro- and meso-fossils (e.g. leaves, flowers and fruits) can have high taxonomic resolution and therefore can be attributed to clades with high confidence. In the case of this well-preserved fossil of Asteraceae, the capitula unambiguously place it outside Asteroideae (Barreda et al., 2012). We used an exponential prior distribution with an offset of 47.5 Myr and a mean of six, which gives a median probability distribution of about 51 Myr that tails off with a 95% probability to 65 Myr. This prior is a broad minimum age constraint that accounts for some uncertainty in the fossil record and corresponds to the most recent common ancestor (MRCA) giving rise to the crown radiation of Asteroideae. A second calibration was placed on the split between the Chatham Island endemics, Olearia chathamica Kirk and O. semidentata Decne. ex Hook.f. This prior corresponds to the emergence of the Chatham Islands. For this calibration point, we followed Wagstaff et al. (2011), but we set a normal prior, with a mean of 3 Mya and a standard deviation of unity. A normal distribution reflects the prior expectation that the most likely divergence is approximately in the centre, but with probable older and younger dates (Ho & Phillips, 2009). Geological evidence suggests that the Chatham Islands may have been completely submerged until 1–3 Mya (Landis et al., 2008). We assumed that the divergence between these two endemic Olearia spp. occurred more recently than this maximum age. However, on the basis of molecular divergence estimates, Heenan et al. (2010) argued that emergent volcanos have persisted in the Chatham archipelago for the last 6 Myr. To avoid the circularity of applying the molecular based estimates of Heenan et al. (2010) as priors in our analysis, we used the younger geological ages, but accommodated error by extending the tails around a normal distribution. Sauquet et al. (2012) demonstrated that estimated ages for nodes nested near the tips of the tree were comparatively more consistent across different calibration scenarios than those of more intermediate ones.
Ancestral area analyses and habitat reconstruction
To explore the major biogeographical events that could explain extant austral disjunct distribution patterns in Lagenophora, we took two approaches: ancestral area analyses and habitat analysis. Distribution, data and habitat descriptions were based on field collections and personal observations, herbarium specimens and pertinent literature (e.g. Cabrera, 1966; Drury, 1974; McQueen, 1976; Wardle et al., 2001; Luebert & Pliscoff, 2006; Kadereit & Jeffrey, 2007 [2006]; Sancho, 2009; Wagstaff et al., 2011). We undertook the biogeographical analysis on the complete set of taxa included in this study, although we focused the results and discussion on the strongly supported Lagenophora core clade, which includes the type species of the genus, L. nudicaulis. The distribution of Lagenophora and the other Astereae was divided into nine areas, based on the presence of one or more endemic species. Some of these areas agree with those defined in other similar biogeographical studies (e.g. Birch & Keeley, 2013). The assigned areas are: A, South Island New Zealand (including Stewart Island); A.1, North Island New Zealand (including Kermadec Islands); B, South Australia (including Tasmania), New Guinea and New Caledonia islands; C, South America [including Falklands (Malvinas) Islands, Juan Fernández Islands, Tristan da Cunha and Gough Island]; D, South-eastern Asia (including surrounding islands); E, Chatham Islands; F, New Zealand Subantarctic Islands (including Auckland and Campbell islands); G, Hawaiian Islands. Outgroup taxa were coded as occurring either in these geographical regions or in South Africa (area H). The area distributions of each species involved in the analysis are indicated in Appendix 1. The Tristan da Cunha archipelago comprises three volcanic islands, which differ in size and age. The largest island is Tristan, being c. 200 000 years old, whereas Nightingale Island is at least 18 million years old. Inaccessible Island is intermediate in age at 3 million years old (Ollier, 1984). To be consistent with the species distribution criterion (i.e. based on the presence of one or more endemic species), and in the absence of Lagenophora samples from the islands, we treated Tristan da Cunha, Gough Island and southern South America as a single unit, although there are geological and historical differences between them. More detailed population studies, beyond the scope of this article, could shed light on the specific relationships between the continent and these remote islands. Preliminary results (not shown) recovered as equally or almost equally probable ancestral areas for a core clade of Lagenophora spp. in the South Island New Zealand (area A) and South Island and North Island of New Zealand (area A–A.1). Because the areas A and A.1 were not informative separately in the preliminary analysis, we used a simpler scheme treating both major islands of New Zealand (South and North Island) as a single unit.
The ancestral area reconstructions were assessed using statistical dispersal–vicariance analysis (S-DIVA) (Yu, Harris & He, 2010) and dispersal–extinction–cladogenesis (DEC) (Ree et al., 2005; Ree & Smith, 2008), both executed in RASP (Reconstruct Ancestral State in Phylogenies) (Yu, Harris & He, 2012). DEC was run including all possible ranges; three unit areas were allowed in ancestral distributions with the possibility to add ranges automatically activated, and the default setting was used for the rest of the parameters. S-DIVA and DEC were run with 10 000 trees obtained by Beast v.1.7.5 from the combined dataset; 1000 samples were discarded before calculating summary statistics and three areas were allowed in the ancestral distributions.
We recognized five types of habitat for Lagenophora in the analysis, mostly based on Wardle's vegetation types (1991): 1. Subalpine tussockland; 2. Beech forest; 3. Coastal dwarf forest; 4. Open scrubland; 5. Coastal dune and cliffs. The habitat analysis was carried out using Fitch parsimony as implemented in Mesquite 2.75 (Maddison & Maddison, 2011).
Results
Phylogenetic analyses
A summary of the levels of variation and tree statistics for the separate and combined DNA matrices is shown in Appendix 2. The number of informative sites in the nrDNA and plastid DNA datasets was typical for these markers in Astereae (e.g. Wagstaff et al., 2011; Bonifacino & Funk, 2012). The incongruence length difference and Shimodaira and Hasegawa tests failed to reveal significant incongruence, and so the data partitions were combined. The MP, ML and Bayesian phylogenetic analyses of the nuclear datasets retrieved quite similar topologies (not shown). The two types of datasets recovered a paraphyletic Lagenophora. Nuclear datasets showed a clade with a core of Lagenophora spp. (LAGC; see Fig. 3 for species) that was strongly supported by BI (ETS, PP = 1; ITS, PP = 1). However, the LAGC clade was not resolved by the plastid markers, perhaps because of the lower number of variable sites or restricted sampling for these markers.
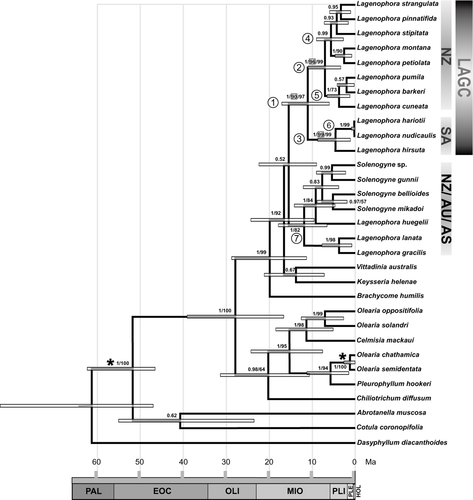
Chronogram from combined nuclear and plastid DNA data modelled under a relaxed clock with uncorrelated rates. Horizontal grey bars on nodes indicate 95% highest posterior density (HPD) of age estimates. Grey scale (0–65.0 Mya) indicates Holocene (HOL), Pleistocene (PLE), Pliocene (PLI), Miocene (MIO), Oligocene (OLI), Eocene (EOC) and Palaeocene (PAL). Posterior probabilities (PP), bootstrap support (BS) for selected clades obtained with maximum likelihood (within squares) and bootstrap support (BS) over 50% obtained with maximum parsimony are indicated above the branches. Numbers of nodes as indicated in Table 2. LAGC, Lagenophora core species; AS, Asia; AU, Australia; NZ, New Zealand; SA, South America. Asterisks indicate the nodes that were constrained for calibration points.
Analyses of the combined dataset by MP, ML and BI in Beast v.1.7.5 resulted in a similar topology (Fig. 3), although MP and ML showed lower internal resolution in the New Zealand clade of Lagenophora. In general, the combined data matrix provided greater resolution and stronger support for relationships than did independent datasets. Notably, the group including LAGC and the L. lanata–Solenogyne clade was unresolved (PP = 0.52; BS less than 50%). However, the L. lanata–Solenogyne clade was recovered as monophyletic with strong support by BI (PP = 1), but weaker support by MP (BS = 82%). Similarly, the LAGC clade was supported by MP, ML and BI (BS = 97%, BS = 90% and PP = 1, respectively). The combined analyses also retrieved two well-supported clades in the Lagenophora core clade. The first comprised New Zealand species (exceptionally extending to Australia): MP BS = 99%; ML BS = 96%; BI PP = 1; within this clade, two monophyletic groups with high support in BI analyses were obtained (PP = 0.99 and PP = 1, respectively). The second clade grouped the South American species (MP BS = 99%; ML BS = 99%; BI PP = 1). The following results will focus mainly on the LAGC clade, which obtained the highest support in the analyses of the combined dataset.
Dating analysis, ancestral area and habitat reconstruction
The values of the substitution rates of each marker obtained in our analyses are shown in Appendix 2. According to our results, substitution rates of nuclear markers (ETS and ITS) were nearly ten times faster than substitution rates of plastid markers (trnK and trnL). The substitution rates for ITS reported here are similar to those reported for some other Asteraceae (e.g. Dendroseris D.Don with values of 0.0039–0.0061, Eupatorium L. with 0.00251, Robinsonia DC. with 0.0079, and tarweeds/Hawaiian silverwords with 0.003; Kay, Whittall & Hodges, 2006; Liu et al., 2006), which provides independent corroboration of the divergence estimates. Variance in nrITS substitution rates outside of Asteraceae has often been reported as both results of errors in the dating process or biological differences among lineages. Thus, distributions of rates should be used with caution to examine specific hypotheses for the timing of events in those plant groups that lack a fossil or biogeographical calibration of their own (Kay et al., 2006).
The estimated divergence times for key nodes in the history of Lagenophora are summarized in Table 2 and Figure 3.
| Node | Age estimate (Mya) | DEC model | S-DIVA model | ||||
|---|---|---|---|---|---|---|---|
| Mean | 95% HPD | 1 | 2 | 1 | 2 | 3 | |
| 1. Lagenophora core | 11.2 | (6.1–17.4) | A|C: 0.64 | A|C: 0.35 | AC: 1.00 | ||
| 2. New Zealand Lagenophora | 7.1 | (3.5–11.2) | A|A: 1.00 | A: 1.00 | |||
| 3. South American Lagenophora | 4.6 | (1.1–8.9) | C|C: 1.00 | C: 1.00 | |||
| 4. New Zealand Lagenophora (I) | 5.7 | (2.8–9.2) | A|A: 1.00 | A: 1.00 | |||
| 5. New Zealand Lagenophora (II) | 3.8 | (1.3–6.7) | A|A: 1.00 | A: 1.00 | |||
| 6. South American Lagenophora (I) | 0.25 | (0–0.83) | C|C: 1.00 | C: 1.00 | |||
| 7. Lagenophora lanata–Solenogyne clade | 12.1 | (6.7–18.2) | A|B: 0.76 | A|A: 0.24 | ABD: 0.33 | AB: 0.33 | B: 0.33 |
Results of the biogeographical analyses are shown in Figure 4 with the probabilities of each ancestral range at the nodes summarized in Table 2. The two biogeographical approaches, DEC and S-DIVA, were largely concordant. Both approaches revealed a complex unresolved origin as the unresolved basal node in Lagenophora. In addition, both approaches supported a New Zealand–southern South America ancestral area (area AC) for the Lagenophora core of species (S-DIVA, 1.00 relative probability; DEC, 0.64 relative probability). The ancestral area for the L. lanata–Solenogyne clade, however, was unresolved for the S-DIVA approach (relative probabilities: area ABD, 0.33; area AB, 0.33; area B, 0.33) and resolved for the DEC approach (area AB, 0.76 relative probability). The ancestral area AC of the Lagenophora core clade suggests a vicariance event that the descendant underwent diversification in the New Zealand area (A) and southern South America area (C). Both approaches, in turn, support dispersal events that led to recent colonization by L. stipitata and L. montana in Australia and Tasmania, and L. pumila and L. petiolata in Subantarctic and Chatham Islands.
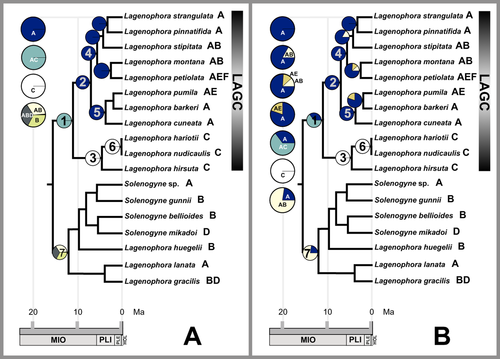
Ancestral area reconstructions on Lagenophora key nodes of Figure 3. A, statistical dispersal–vicariance analysis (S-DIVA) ancestral area reconstruction; colour codes when possible as in Figure 1. B, dispersal–extinction–cladogenesis (DEC) ancestral area reconstruction. Circles represent ancestral areas. Area distributions in the figure: A, New Zealand; B, South-eastern Australia, New Guinea and New Caledonia; C, southern South America; D, South-eastern Asia (including surrounding islands); E, Chatham Islands; F, Subantarctic Islands. Numbers in circles indicate number of nodes as in Figure 3 and Table 2; bold capital letters beside the species names indicate area distributions (for a complete list, see Material and Methods). LAGC, Lagenophora core species. Grey scale as in Figure 3.
Figure 5 shows the habitat reconstruction for the Lagenophora core clade whose basal node, in turn, recovered a congruent ancestral area under both biogeographical approaches. According to the habitat reconstruction, beech forest is the putative ancestral habitat for the South American clade, but with ambiguous results for the New Zealand clade with both beech forest and tussockland as ancestral habitats. Colonization of the other habitats occurred independently in different lineages.
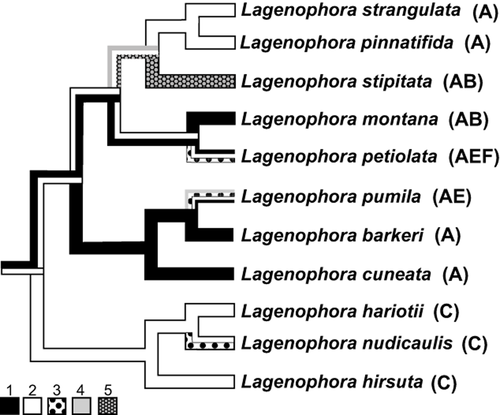
Habitat reconstruction on Lagenophora core clade: 1, Subalpine tussockland; 2, Beech forest; 3, Coastal dwarf forest; 4, Open scrubland; 5, Coastal dune and cliffs. Bold capital letters beside the species names indicate area distributions. Area distributions in the figure: A, New Zealand; B, South-eastern Australia, New Guinea and New Caledonia; C, southern South America; E, Chatham Islands; F, Subantarctic Islands.
Discussion
Our results suggest that Lagenophora is a recent lineage that diversified long after the break-up of Gondwana. As currently circumscribed, Lagenophora is not monophyletic, but we identify two smaller clades that are well supported: the Lagenophora core clade (including the type species of the genus) and the L. lanata–Solenogyne clade. We consider alternative scenarios based on molecular dating and biogeographical reconstructions. The initial radiation of Lagenophora began in the mid-Miocene and continued in the late Miocene–early Pliocene, giving rise to geographically isolated clades in Australia, New Zealand and South America. The S-DIVA and DEC biogeographical reconstructions were largely congruent for the Lagenophora core clade and recovered a complex ancestral area which, with the estimated divergence times and habitat reconstructions, provides circumstantial evidence that Antarctica could have served as a corridor for overland migration until the expansion of the continental ice forced these lineages to migrate northwards during the late Cenozoic. The crown radiations in the geographically isolated lineages in Australia, New Zealand and South America occurred during the late Pliocene/Pleistocene, which suggests that geological and climatic changes at this time may have been important drivers of speciation.
Phylogeny of Lagenophora and taxonomic implications
According to our results, most Lagenophora spp. grouped in a well-supported clade which we called the Lagenophora core. The Australian and Asian species L. huegelii, L. gracilis and L. lanata, however, were recovered in a monophyletic group (Fig. 3, node 7) with Solenogyne spp., in agreement with Nakamura et al. (2012, based on ITS). Contrary to these authors, we obtained weak support for the relationships between these two clades. These differences could be a result of the more diverse outgroups and hence greater homoplasy in our approach. Our studies, therefore, do not strongly support a broad sense of Lagenophora, which, in turn, would be morphologically remarkably diverse. Alternatively, a narrow circumscription of Lagenophora involving only the species of our Lagenophora core clade would reflect more clearly its biogeographical and evolutionary history. However, a final taxonomic decision on the circumscription of Lagenophora requires further investigation involving other markers for all species of this genus and Solenogyne, and a deep morphological perspective of the group.
Role of fruits in Lagenophora dispersibility
Our results show that at least part of the biogeographical history of Lagenophora has been influenced by dispersal events, which could be facilitated by its unusual floral features. The pappus and the typical fruit trichome of the family, thought to prevent desiccation, are lacking in Lagenophora. The fruits are covered by glands with a sticky secretion (Fig. 2F). An umbrella-like ‘infructescence’ (Fig. 2G) emerges from the otherwise short plants usually embedded in other understorey plants or mosses, and exposes the sticky fruits. According to Carlquist (1967, 1983), viscid fruits are suitable for attachment to birds. Also, the possibility that birds could have been dispersal vectors for Lagenophora fruits was postulated by Cabrera (1966) to explain some of the extant distributions of the genus. Alternatively, successful overwater dispersal events could be postulated for Lagenophora, if the oily fruit cover is suggested to aid in flotation, as was the case for other families (Prance & Mori, 1979). Thus, the absence of pappus in Lagenophora would not result in the absence of dispersal mechanisms. On the contrary, the sticky fruits, with the ‘infructescence’, apparently act as an efficient dispersal mechanism involving several potential dispersal vectors.
Antarctica as a suitable area for Lagenophora ancestors
Although with differences in the timing of events, our results lend support to the scenario proposed by Wagstaff et al. (2006) for Abrotanella, indicating that Antarctica may have played a key role as a corridor for migration between the austral landmasses. New Zealand had rafted away from Antarctica earlier than the calculated divergence time of Lagenophora; thus, events related to the break-up of Gondwana are unlikely. Our S-DIVA and DEC analyses placed the ancestral area of the Lagenophora core in a complex area historically encompassing New Zealand and South America. It has been suggested that Antarctica was a corridor for migration until the late Cenozoic, when expanding ice sheets completely eliminated the flora (c. 3.9–14 Mya; Axelrod, Arroyo & Raven, 1991; Ashworth & Cantrill, 2004; Convey et al., 2008; Lewis et al., 2008; Benedetto, 2012). It could be the case that ancestors of Lagenophora first migrated along the Antarctic coast, which would agree with other studies undertaken in Asteraceae (Wagstaff et al., 2006). Indeed, it has been suggested that birds bridged oceanic gaps between Antarctica, New Zealand, Tasmania and South America during the mid-Miocene warm interval, facilitating transportation of small plants and animals (Ashworth et al., 2007). This kind of transportation could have involved Lagenophora sticky fruits as indicated by some authors (Cabrera, 1966; Carlquist, 1967). The estimated age of the split between the New Zealand and South American Lagenophora spp. is 11.2 Mya [6.1–17.4 95% highest posterior density (HPD)]. The Antarctic coastal areas and Transantarctic Mountains supported beech-dominated habitats at this time (Axelrod et al., 1991; Swenson & Bremer, 1997; Warny et al., 2009). In addition to good dispersal abilities, successful colonization requires both the availability of habitat and an ability to establish; it could be possible that Antarctic suitable beech-dominated habitats facilitated the gene flow of Lagenophora ancestors between southern South America and New Zealand. In this scenario, one could postulate a relatively continuous distribution along the Antarctic coastline for Lagenophora ancestors. The connections via Antarctica could have been interrupted when the ice sheet completely covered its surface in the middle Miocene–Pliocene boundaries. By that time, the estimated divergence of the New Zealand and South American Lagenophora groups could have occurred, thus suggesting gene flow restriction between both lineages.
Recent speciation of Lagenophora in New Zealand
Our results indicate that the New Zealand clade diversified at 7.1 Mya (3.5–11.2 95% HPD) in the late Miocene–early Pliocene. The ancestors of Lagenophora fall into two clades that diverged at 5.7 Mya (2.8–9.2 95% HPD) and 3.8 Mya (1.3–6.7 95% HPD). Lagenophora is an element of the cool temperate New Zealand flora that developed during the Miocene as part of a southern extension of the New Zealand archipelago (Wardle, 1963). This flora gave rise to the present mountain flora after the onset of orogeny and climatic cooling in the Pliocene. However, Fleming (1963) favoured Antarctica as a potential refuge for cool-adapted lineages. The increasing tectonic activity of the Pliocene that led to the rapid elevation of the axial mountains of New Zealand (Winkworth et al., 2005), and glaciations during the late Pliocene and Pleistocene, could have driven diversification in Lagenophora by habitat diversification, as has been postulated for other lineages (e.g. Wardle, 1968; Wagstaff & Breitwieser, 2004). Indeed, Lagenophora spp. in New Zealand are found in diverse environments. Some New Zealand Lagenophora spp. are restricted to specific environments, such as those provided by light gaps in the dense beech forests (e.g. L. strangulata or L. pinnatifida) or by the subalpine tussockland (i.e. L. barkeri, L. cuneata and L. montana). Others, such as L. pumila and L. petiolata, colonized diverse environments in the Chatham Islands and the Subantarctic Islands.
The MP reconstruction of ancestral habitats indicates that Lagenophora ancestors in New Zealand initially inhabited light gaps and margins in beech forest, as well as subalpine tussockland communities, and from there diversified into other diverse environments. The coastal areas of New Zealand Subantarctic and Chatham Islands were colonized at least twice independently, and these events occurred within approximately the last 2 Myr. Thus, our results suggest a recent colonization of New Zealand Subantarctic Islands instead of viewing the extant distribution of Lagenophora in these areas as a remnant of an older biota (Wardle, 1963). Pleistocene biotic exchange through the Chatham Rise, a geological connection between the South Island of New Zealand (east coast) and the Chatham Islands, has been suggested by Fleming (1979) and Heenan et al. (2010).
Trans-Tasman distribution of Lagenophora
The historical biogeography of Lagenophora in New Zealand indicates that range expansions to new regions occurred sporadically in different phylogenetic lineages. It has been suggested that lineages inhabiting the current mountains of Australia and New Guinea arose following dispersal from New Zealand (Winkworth et al., 2002). Other microfossil studies (Truswell & Macphail, 2009) also support eastward dispersals. Specifically, for Solenogyne [according to Nakamura et al. (2012) closely related to Lagenophora], colonization of Asian archipelagoes from Australia followed either long-distance dispersal or extinction of intermediate populations. Our Lagenophora core clade biogeographical reconstruction would agree with the scenario of recent dispersal events from New Zealand to Australia and Asia. However, these events would be confirmed by specific population studies.
Recent speciation of Lagenophora in southern South America
The results of our habitat reconstruction show beech forest as the ancestral habitat for the South American clade of Lagenophora, supporting the association between both taxa. According to our analyses, the presence of Lagenophora in South America dates from about 4.6 Mya (1.1–8.9 95% HPD), suggesting that its ancestors had been in this continent by the late Miocene–Pliocene. Fossil records placed beech-dominated tundra-like floras in Antarctica by the middle to late Pliocene (Francis & Hill, 1996; Ashworth et al., 2007; Warny et al., 2009). If these records are certain, beech-dominated habitats could have facilitated the settlement of Lagenophora ancestors throughout Antarctic coastlines. Indeed, it has been postulated that Neogene refugia were present on the margins of Antarctica, from which plants and animals could migrate to the continental interior during a time or times of significant climatic warming (Haywood et al., 2009). It has been postulated that phylogenetic biome conservatism had a prevalent role during the radiation of plant lineages of southern landmasses, both within continents and in trans-oceanic colonization (Crisp et al., 2009), which could be the case for Lagenophora in association with beech-dominated habitats.
In southern South America, during the Miocene, the largest Andean uplift and the increasing cooling led to extremely arid conditions in eastern Patagonia, which resulted in a deep differentiation of Andean and extra-Andean environments. This new environmental condition in the Miocene restricted the beech forest almost exclusively to the Andean region. During the Pliocene and Pleistocene, glacial periods and the new rain gradient resulting from the Andean uplift pushed the cool temperate forest to its current Andean domain (Iglesias, Artabe & Morel, 2011). Studies on Antarctic Nothofagaceae (Francis & Hill, 1996) during the Pliocene suggested a low prostrate habit, more similar to shrubs growing today along exposed parts of the treeline on Isla Navarino in southernmost Chile. Nowadays, beech forests in southern South America are restricted to median to high elevations (up to 2000 m) in the southernmost Andes from c. 33°S to the southernmost extremes of Argentina and Chile (McQueen, 1976; Luebert & Pliscoff, 2006; Moreira-Muñoz & Muñoz-Schick, 2007). The reconstruction of the historical biogeography of Lagenophora, as presented here, in the frame of the apparent association of this genus with the southern cool-temperate beech forest, could contribute to our understanding of the spatio-temporal patterns of the austral biota.
According to our spatio-temporal and habitat reconstructions, Lagenophora ancestors could have survived the extreme aridity that predominated in the Andean regions after the Andean uplift in the Miocene, followed by retraction of the beech forest to these mountains. Although a Pliocene crown age for the South American clade was inferred, differentiation of the two most recent species of Lagenophora (L. hariotii and L. nudicaulis) dates from the Upper Pleistocene, 250 Kya (0–830 95% HPD), when climatic fluctuations drastically affected the South American biota. During the Last Glacial Maximum (LGM), c. 20 Kya, the Andes were covered by icefields. According to Fraser et al. (2012), numerous cold-resistant taxa survived in glacial and periglacial refugia east of the Andes and Tierra del Fuego with expansion to high latitudes following glacial periods. These environmental changes probably influenced the extant distribution of Lagenophora in South America. Nowadays, the three southern South American Lagenophora spp. are restricted to the beech forest understorey. They inhabit light gaps and borders of the forest. Lagenophora nudicaulis and, exceptionally, L. hariotii were capable of colonizing new habitats (Fig. 2D), once settlement in beech forest by the ancestors of Lagenophora in South America was accomplished. Lagenophora nudicaulis has dispersed widely to island archipelagoes, such as the Falklands and the remote Tristan da Cunha and Gough Island. Cabrera (1966) explained this geographical distribution by transportation of Lagenophora fruits by petrels and shearwaters. Alternatively, during the LGM, the sea level fell 120 m extending the Patagonian coast 300 km eastwards (Benedetto, 2012). This could have aided the expansion of L. nudicaulis from the continent towards the Falkland Islands.
Recent uplift of the axial mountains of Australia, New Zealand and the Andes of southern South America created new habitats, dispersal routes and barriers that probably played a key role in the diversification and settlement of Lagenophora. Climatic changes in the Neogene also apparently affected the southern biota. Cooling could have had more influence in New Zealand, whereas cooling and extreme aridity have dramatically affected the flora of southern South America (Iglesias et al., 2011; Benedetto, 2012). Whilst the colonization of new ecological niches apparently dominated diversification of Lagenophora in New Zealand, biome conservatism appears to have been the more likely hypothesis for this genus in South America. Probably, a unique scenario cannot explain the extant austral distribution of Lagenophora; instead, it could be a result of a complex history involving different dispersals, and recent vicariance events may be related to the establishment of the Antarctic ice sheet. In any case, the connection between the ancestors of Lagenophora of both New Zealand and southern South America was apparently interrupted by the mid-Miocene–early Pliocene [11.2 Mya (6.1–17.4)], leading to diversification in New Zealand separately from diversification in South America.
Acknowledgements
This work was funded by Agencia Nacional de Promoción Científica y Tecnológica (Secretaría de Ciencia y Técnica PICT 2007-01977), Comisión Nacional de Investigaciones Científicas y Técnicas, Argentina to GS and the Royal Society of New Zealand (2007–08 ISAT Linkages Fund) to GS. We acknowledge Rob Smissen, Leon Perrie and the reviewers for useful comments on the manuscript, Tom Ranker for providing the Keysseria sample, Peter Fritsch and Mauricio Bonifacino for their photographs in Figure 2, Mario Aguilar and the IBBM laboratory staff Universidad de La Plata (UNLP) (GS) and Nicola Bolstridge (Landcare Research) for assistance in the laboratory.
Appendix 1
Voucher information, GenBank accession numbers for sequences used in this study and area distributions. Voucher information listed in the following order: taxon name, country of origin, locality, collection, herbarium, reference (if sequence taken from GenBank). GenBank numbers listed in the following order: ITS, ETS, trnK, trnL. MS, missing sequence. Capital letters in parentheses indicate area distributions.
Abrotanella muscosa Kirk, New Zealand, Stewart Island, Breitwieser 2116 and Wilton, CHR 534876, Wagstaff et al. (2011), AF422109, HQ439820, AY554052, HQ439865, (A). Brachycome humilis G. Simpson and J. S. Thomson, New Zealand, cult. ex South Island, P. Heenan s.n., CHR 514150, Wagstaff et al. (2011), AF422113, HQ439822, HQ439791, HQ439867, (A). Celmisia mackaui Raoul, New Zealand, cult. ex South Island, Canterbury, Wardle and MacRae s.n., CHR 514149, Wagstaff et al. (2011), AF422115, HQ439825, HQ439792, HQ439870, (A). Chiliotrichum diffusum (G. Forst.) Kuntze, Argentina, Río Negro, Parque Nacional Nahuel Huapi, Ezcurra 2376, CHR 530116, Wagstaff et al. (2011), AF422117, HQ439827, HQ439794, HQ439872, (C). Dasyphyllum diacanthoides (Less.) Cabrera, Argentina, Río Negro, Bariloche, Wardle and Wagstaff 97120, CHR 514092, Wagstaff et al. (2011), AF422120, HQ439830, AY554072, HQ439875, (C). Keysseria helenae (C.N. Forbes and Lydgate) Cabrera, USA, Hawaii, Sincock Bog, Wood 12265, HI, KP017344, KP017335, KP017320, KP017358, (G). Lagenophora barkeri Kirk, New Zealand, Lake Tennyson, de Lange 7278, CHR 658552, KP017349; MS; New Zealand, without data, CHR 309607, KP017325, KP017360, (A). Lagenophora cuneata Petrie, without data, Gardner et al. (2007, New Zealand biodiversity database), EU352246; New Zealand, Porters Pass, Wagstaff and Sancho s.n., CHR 605144, KP017326, KP017312, KP017350, (A). Lagenophora gracilis Steetz, Australia, Queensland, NK 20100013, RYU, Nakamura et al. (2012), AB550254; MS, MS, MS, (B, D). Lagenophora hariotii Franch., Argentina, Tierra del Fuego, Bonifacino et al. 3020a, LP, KP017342, KP017333, KP017318; Bonifacino et al. 3020b, LP, KP017356, (C). Lagenophora hirsuta Poepp. ex Less., Argentina Río Negro, Sancho and Bush 109, LP, KP017343, KP017334, KP017319, KP017357, (C). Lagenophora huegelii Benth., Australia, Victoria. NK20100014, RYU, Nakamura et al. (2012), AB550255; MS, MS, MS, (B). Lagenophora lanata A. Cunn., New Zealand, North Island, Te Paki Ecological Region, de Lange 10274, CHR 10274, KP017345, KP017336, KP017321; MS, (A). Lagenophora montana Hook. f., without data, Gardner et al. (2007, New Zealand biodiversity database), EU352243; New Zealand, Lake Sara, Wagstaff and Sancho s.n., CHR 605087, KP017327, KP017313; MS, (A, B). Lagenophora nudicaulis (Comm. ex Lam.) Dusén, Argentina, Tierra del Fuego, Sancho and Plos 184, LP, KP017347, KP017338, KP017323; MS, (C). Lagenophora petiolata Hook.f., New Zealand, Franz Josef Glacier, Sancho et al. 104, CHR 605102, KP017340; New Zealand, Boyle River, Wagstaff and Sancho s.n., CHR 605107, KP017328; New Zealand, Campbell Islands, Rance 7279, CHR 605090, KP017314, KP017351, (A, E, F). Lagenophora pinnatifida Hook. f., New Zealand, Mt Arthur, de Lange s.n., AK, KP017341, KP017329, KP017315, KP017352, (A). Lagenophora pumila (G. Forst.) Cheesman, New Zealand, cult. ex South Island, Heenan s.n., CHR 514151, Wagstaff et al. (2011), AF422124 (ITS), HQ439832 (trnK); New Zealand, without data, CHR 423718, KP017330 (ETS); New Zealand, Boyle River, Wagstaff and Sancho s.n., CHR 605106, KP017353 (trnL), (A, E). Lagenophora stipitata (Labill.) Druce, without data, Shimamura and Watanabe (2009, published only in database), AB435145; New Zealand, Woodhills Forest, de Lange 7276, CHR 605089, KP017331, KP017316, KP017354, (A, B). Lagenophora strangulata Colenso, New Zealand, without data, Gardner et al. (2007, New Zealand biodiversity database), EU352245; New Zealand, Lyndon Pass trail, Wagstaff and Sancho s.n., CHR 605143, KP017332, KP017317, KP017355, (A). Olearia chathamica Kirk, New Zealand, cult. DOC Nursery, Motukarara, ex Chatham Islands, Wagstaff s.n., CHR 529989, Wagstaff et al. (2011), AF422129, HQ439835, HQ439799, HQ439880, (E). Olearia oppositifolia (F. Muell.) Lander, Australia, New South Wales, Barrington tops, UNSW 24149, Cross et al. (2002), AF497709, HQ439844, HQ439808, HQ439889, (B). Olearia semidentata Decne. ex Hook. f., New Zealand, Chatham Island, de Lange CH334, CHR 566555, Wagstaff et al. (2011), HQ439862, HQ439848, HQ439812, HQ439893, (E). Olearia solandri (Hook.f.) Hook.f., New Zealand, cult., UNSW 24099, Cross et al. (2002), AF497696, HQ439849, HQ439813, HQ439894, (A). Pleurophyllum hookeri Buchanan, New Zealand, Campbell Island, Meurk s.n, CHR 537467, Wagstaff et al. (2011), HQ439864, HQ439853, HQ439917, HQ439898, (F). Solenogyne bellioides Cass. Australia, New South Wales, NK20100010b, Nakamura et al. (2012), AB604756.1; MS, AB543928.1, MS, (B). Solenogyne gunnii (Hook.f.) Cabrera, New Zealand, North Island, Waikato, cult., de Lange 10299, AK, KP017346, KP017337, KP017322, KP017359, (B). Solenogyne mikadoi Koidz., Japan, Iriomotejima Island, without data, NK20100009, Nakamura et al. (2012), AB543934.1; MS; Japan, Okinawajima Island, NK20100004, Nakamura et al. (2012), AB543926.1, MS, (D). Solenogyne sp., New Zealand, cult. ex Clutha River, Otago, South Island, Barkla s.n., CHR 605697, KP017348, KP017339, KP017324; MS, (A). Vittadinia australis A. Rich., New Zealand, Banks Peninsula, Sykes 29/95, CHR 500653, Wagstaff et al. (2011), AF422140, HQ439856, HQ439819, HQ439901, (A).
Appendix 2
Selected models used in maximum likelihood and Bayesian analyses and summary of tree statistics analysed under parsimony criteria for separate and combined DNA matrices; results of incongruence length difference (ILD) and Shimodaira and Hasegawa (SH) tests; substitution rates per site per million years obtained in the log files created by our BEAST analyses. CI, consistency index; ETS, external transcribed spacer; HPD, highest posterior density; ITS, internal transcribed spacer; MPTs, number of trees obtained by maximum parsimony; RI, retention index.
| Bayesian analysis: selected model | Aligned length | Informative characters | MPTs | Tree length | CI | RI | ILD | SH | Substitution rate | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | 95% HPD | ||||||||||
| ETS | TIM2 + G | 423 | 144 | 12 | 520 | 0.70 | 0.71 | – | – | 0.00497 | 0.00356–0.00637 |
| ITS | SYM + G | 693 | 150 | 5 | 576 | 0.74 | 0.75 | – | – | 0.00298 | 0.00211–0,00380 |
| trnK | TIM1 + I + G | 868 | 35 | 1 | 132 | 0.90 | 0.91 | – | – | 0.00040 | 0.00026–0.00052 |
| trnL | TPM3uf + G | 832 | 34 | 1 | 130 | 0.93 | 0.92 | – | – | 0.00052 | 0.00035–0.00067 |
| Combined | TIM1 + I + G | 2822 | 363 | 9 | 1385 | 0.74 | 0.73 | 0.73 | 0.54 | ||




