Heritabilities for copy number variation of porcine endogenous retrovirus by a quantitative PCR
Abstract
Pigs (Sus scrofa) have been expected to have organs transplanted to humans, but porcine endogenous retrovirus (PERV) is one of the risks because the PERV has the possibility to get infected with human cells. Therefore, the pigs are required to have as low a PERV copy number as possible. In this study, firstly, we investigated the estimates of heritabilities for the PERV copy numbers in the Vietnamese native breeds. Genomic heritabilities for four genes on PERV were estimated using the restricted maximum likelihood method with the genomic relationship matrix. The genomic heritability estimates of these genes ranged from 0.27 to 0.71, indicating that it would be possible for these genes not to follow the normal Mendelian inheritance. Secondly, we bred the pig population to reduce the pol gene number and estimated the heritability for the number. Despite the high heritability estimate for the pol gene (0.59), little improvement was progressed after selection for reducing the gene number in the three generations. In order to reduce the PERV copy numbers from the pig genome, it would be difficult to adapt only conventional breeding technology, and we need to consider using another technology like genome editing.
1 INTRODUCTION
Variations in the mammalian genome have been found from single nucleotide variants or polymorphisms (SNPs) to large insertions or deletions. A contiguous block of DNA sequence is rarely duplicated by DNA duplication mistakes, which are called a copy number variation of a gene. In pig breeds, KIT and MC1R are familiar with duplicated genes (Giuffra et al., 2002; Kijas et al., 1998), and the variations are related to coat colors in many breeds. In the human genetic study, Locke et al. (2006) showed that heritability estimates for most of the copy number variations were almost one and consistent with normal Mendelian inheritance because these loci are in strong linkage disequilibrium. In another case of copy number variations, there are a lot of virus-derived sequences on the pig genome and transposable elements like short interspersed nuclear elements (SINEs) and long interspersed nuclear elements (LINEs) on genomes (Groenen et al., 2012). These sequences are endowed with the ability to transpose themselves into another locus. Particularly, porcine endogenous retrovirus (PERV), which is a provirus integrated into the pig genome, is found in genome-wide regions. The virus sequences could be expected to be inherited from parents to offspring according to Mendelian inheritance (Weiss, 2016). However, as far as we know, no referential result has been shown that PERV follows Mendelian inheritance.
Organs from domestic pigs have been expected to be xenotransplants of humans. For one of the risks, PERV may act as an infectious source causing unexpected disease in humans during/after xenotransplantation exists (Magre et al., 2003). Previous studies have not reported a clear relationship between the number of PERV genes on the genome and the occurrence of PERV transmission in human cells. However, from a perspective of fewer risk factors, Yang et al. (2015) and Niu et al. (2017) attempted to adopt a CRISPR-Cas9 technology for domestic pigs in order to avoid any transmission of PERV genes. However, the CRISPR-Cas9 technology frequently causes mutations on off-target, which may cause genomic instability and disrupt the functionality of otherwise normal genes, and it remains still one major concern when applying the CRISPR/Cas9 system to biomedical and clinical applications. In addition, even if genome editing technology applies to genes that have many copies on the genome-wide location, the high mortality ratio of embryos becomes the next issue. For these reasons, it is primary to find or develop pigs with a lower PERV copy number. An alternative approach is to use the conventional animal breeding approach to reduce or eliminate the copy of the PERV in the pig population. In this approach, we treat the number of copies of the PERV genes as a phenotype, and pigs with the low number in each generation are selected as parents of the next generation.
Different pig breeds have different copy numbers of PERVs (Denner, 2016), and Western domestic breeds contain many copies, as previous studies showed (Ishihara et al., 2020; Liu et al., 2011; Mang et al., 2001). In contrast, we showed that the copy number of the PERV gene was significantly lower in Vietnamese native pig breeds compared with Western breeds in a previous study (Ishihara et al., 2020). Many Vietnamese native pig breeds are classified as mini pigs because of their small body size. Minipig breeds have characteristics that make them ideal as xenotransplant material, as their organs are similar in size to humans.
Here, we investigated the genetic characteristics of the PERV gene in the Vietnamese native pig breeds. We first investigated how the amount of genomic differentiation could explain the differentiation of the copy number of PERV genes. Secondly, we estimated the heritability of the copy number of a PERV gene and predicted its selection response using a conventional breeding technology. Our results provide some key parameters needed for developing pigs with lower PERV copy numbers for medical purposes.
2 MATERIALS AND METHODS
Our study consists of two experiments. The first experiment is that we attempted to estimate genomic heritabilities for copy numbers of the genes on PERV by using the data of Ishihara et al. (2018) and Ishihara et al. (2020). Genomic heritability can be regarded as the proportion of variance of phenotypes that is accounted for variation captured by SNPs linked with PERV genes (de Los Campos et al., 2015). The data from the first experiment have already been published (Ishihara et al., 2018, 2020), and thus, the first experiment of this study was just carried out to estimate genomic parameters using the published data.
In the second experiment, a new experimental family was constructed, and a breeding program to reduce the copy number was implemented. According to the previous survey (Ishihara et al., 2020), we identified the region where the Vietnamese native pig breed would have the low PERV copy number, and the base animals of the family were brought from the region. In this experiment, we show the result of the breeding program for the experimental family to reduce the copy number of the pol gene on PERV. The present animal study protocol followed the Guidelines of Animal Care and Use of National Institute of Animal Science (NIAS), Vietnam, and the data of the phenotypic and pedigree of the experimental family are provided as Supporting Information.
3 THE FIRST EXPERIMENT
3.1 Data preparation
The first experiment is based on the data from Ishihara et al. (2018) and Ishihara et al. (2020). The samples consisted of six individuals sampled from each of the 15 breeds based on a report from the Vietnamese Ministry of Agriculture and Rural Development (Tạ et al., 2013). A total of 90 pigs were used to estimate genomic heritabilities for the copy numbers of the genes on PERV. The details of the data were written in Ishihara et al. (2018) and Ishihara et al. (2020). The plink software of v1.09 (Purcell et al., 2007) was used for the quality check. We only used SNPs on the autosomal chromosomes. The SNPs with a minor allele frequency (MAF) lower than 0.05, a missing call rate lower than 0.10, and a p-value of Hardy–Weinberg equilibrium lower than 0.01 were removed from the. After quality control, 32,863 SNPs were used for estimating genomic parameters.
3.2 Genomic analysis
4 THE SECOND EXPERIMENT
4.1 Animals and DNA preparation
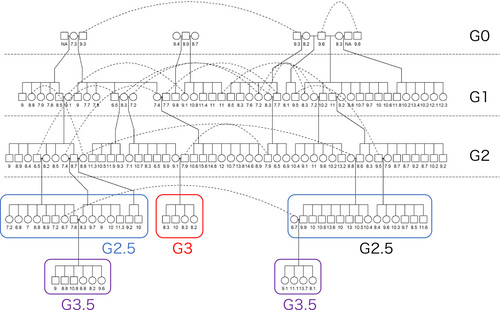
The second experiment is that we attempted to reduce the numbers of the pol gene using the conventional breeding design. The pol gene encodes a reverse polymerase, which is the catalytic core among PERV A, B, and C, and because the pol gene is common for the three PERV, we can estimate the total number of the PERV copies on the genome. In the previous study on genome editing for PERV, the pol gene was targeted to inactivate the PERV in cells (Niu et al., 2017). The base animals in this population were collected from Yen Bai province, Vietnam, according to the previous survey (Ishihara et al., 2020). Those animals were selected as far as they could be ascertained for relationships between pedigrees, based on interviews with the farmers concerned, and were taken to the breeding facility of NIAS in Thai Nguyen province, Vietnam. We adopted a closed population scheme, a so-called simple animal breeding scheme, to reduce the population average of the copy number of PERV. In this scheme, we treated a measurement of the copy number of PERV by the real-time PCR. In each generation, animals at each generation that have as a low number of PERV as possible were selected to be parents of the next generation, and then these animals were mated to produce progenies as a candidate for the next parents. We planned the discrete generation mating, which means that selection was conducted on each generation, and only selected animals were kept as parents. The other animals that had high values of PERV copy number were culled after we measured the copy number of PERV. Unfortunately, in the second generation, we were not able to obtain a sufficient number of offsprings to be selected for the next generation, and thus, temporarily, we mated animals belonging to the first generation with ones of the second generation, which are treated as the 2.5 generation (see Figure 1). Although we proceeded to produce the next generation (3.5 generation), these progenies had a relatively high PERV copy number, and thus no animal belonging to the 2.5 and 3.5 generations was finally selected as the candidate. Therefore, the current population consists of only the animals belonging to the third generation. All pigs have been identified by an ear tag after approximately 10 days at birth. We collected the ear tissues at the same time, and genomic DNA was extracted from these tissue samples using the QiAmp DNA mini kit (QIAGEN Ltd., Hilden, Germany) in accordance with the manufacturer's instructions.
4.2 Quantitative analysis of PERV copy number
The copy numbers of the pol gene on PERV were estimated by the method using absolute quantification of the TaqMan® real-time PCR system (Applied Biosystems). Briefly, we used β-actin (ACTB) as the best stable internal control gene to be considered a single copy in the genome. The real-time PCR mixtures for DNA amplification contained 7.5 μL of 2× TaqMan gene expression master mix (Applied Biosystems), 10 μM of both forward and reverse primers and 5 μM of probes for ACTB and the target genes, 5 ng DNA of each individual, and distilled water to a final volume of 15 μL. The PCR program consisted of a denaturation step at 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All reactions were performed in triplicate. A dissociation curve analysis was performed to verify the specificity of the amplified products. Quantitation of ACTB and the target genes was performed using the standard curve created using plasmid DNA clones for ACTB and the target genes. Therefore, in this study, we defined the unit of copy number of the pol gene in the pig genome as follows:
the pol gene copy number on PERV = Absolute quantification (the pol gene)/Absolute quantification (ACTB)
4.3 Pedigree analysis
Before estimating genetic parameters, an ANOVA with fixed effects was performed using the R software (R Core Team, 2020). The effects in the model were sexes and generations. Statistical significance was considered at a p-value < 0.05.
We also used the REML with the average information to estimate variance components.
5 RESULTS
5.1 The genomic heritabilities for copy numbers of the genes on PERV
According to Ishihara et al. (2020), the average copy numbers of gag, pol, envA/C, and envB on PERV were 19.48 ± 5.12, 16.97 ± 4.62, 9.80 ± 7.65, and 6.82 ± 2.23, respectively. Ishihara et al. (2020) also showed that the differences in PERV copy numbers were found in the population we analyzed, and the Mong Cai and Ba Xuyen breeds showed significantly higher values on average compared with the other Vietnamese breeds. The histograms for the copy numbers are shown in Figures S1–S4, and the copy numbers of all genes deviated from normality. Heritability estimates for the traits and their standard error are presented in Table 1. We obtained the moderate (0.267 ± 0.388 for envB) to high heritability estimates (0.634 ± 0.290 and 0.708 ± 0.309 for gag and envA.C, respectively). However, in all traits, the standard errors of the heritabilities were quite high, with values of approximately 0.3.
| Parameter | gag | pol | envA.C | envB |
|---|---|---|---|---|
| Genomic variance | 11.274 ± 6.189 | 4.210 ± 3.850 | 0.094 ± 0.051 | 0.753 ± 1.156 |
| Residual variance | 6.498 ± 4.696 | 6.313 ± 3.112 | 0.039 ± 0.037 | 2.066 ± 0.959 |
| Heritability | 0.634 ± 0.290 | 0.400 ± 0.333 | 0.708 ± 0.309 | 0.267 ± 0.388 |
The correlations in phenotypic and genomic levels for four PERV genes are presented in Table 2. The correlations between the three genes of gag, pol, and envA.C were quite high, and the values were more than 0.5 in both phenotypic values and genomic breeding values, except for envB. Meanwhile, the envB gene has less correlation with the other three genes, and we obtained negative values for the correlations in the phenotypic and genomic levels between envA.C and envB.
| Genes | Genes | |||
|---|---|---|---|---|
| gag | pol | envA.C | envB | |
| gag | 0.796 | 0.623 | 0.238 | |
| pol | 0.741 | 0.541 | 0.321 | |
| envA.C | 0.602 | 0.716 | −0.087 | |
| envB | 0.150 | 0.167 | −0.246 | |
5.2 Phenotypic variation and heritability in the breeding population
In the second experiment, PERV copy number was measured in a total of 134 pigs (Table 3). In this population, the average measurement in the base population was 8.706 ± 0.581. The histogram for the pol gene number is shown in Figure S5, which shows that the copy number of the gene slightly deviated from normality. The ANOVA test showed that the copy number of the pol gene differed significantly between sexes (p < 0.05). Although we obtained the pig with a smaller copy number (6.465) in the first generation than the minimum value in the base population (7.300), the average value in the first generation was about one point higher than one in the base population (9.667 ± 2.397). After the first generation, the averages were gradually decreasing, and in the third generation, although the samples are quite small (two males and two females), the average was similar to one of the base population (8.695 ± 0.759). However, there was no change between the maximum and maximum values of the copy number of pol during the generations.
| Generation | Number of animals | Mean | Standard deviation | Maximum | Minimum | |
|---|---|---|---|---|---|---|
| Male | Female | |||||
| 0 (base) | 3 | 5 | 8.706 | 0.581 | 9.590 | 7.300 |
| 1 | 16 | 16 | 9.667 | 2.397 | 13.400 | 6.465 |
| 2 | 25 | 22 | 9.567 | 4.680 | 15.611 | 6.472 |
| 2.5 | 16 | 13 | 9.387 | 3.172 | 13.570 | 6.691 |
| 3 | 2 | 2 | 8.695 | 0.759 | 9.997 | 8.160 |
| 3.5 | 5 | 9 | 9.516 | 3.772 | 13.698 | 6.773 |
The estimates and standard errors of genetic and residual variance components and heritability for the copy number of pol were 2.033 ± 0.739, 1.389 ± 0.412, and 0.594 ± 0.074, respectively. The estimate of heritability obtained in the breeding population was higher than that in the first experiment (0.400 ± 0.333).
6 DISCUSSION
In this study, we attempted to estimate genetic parameters for copy number variation of PERV genes using the whole breeds in Vietnam and data from the breeding herd. Gene duplication, leading to gene copy number variants in the population, generally happens during crossing-over. Therefore, duplicate genes are generally located in close regions where the original gene is located, and thus they are in tight linkage. Locke et al. (2006) attempted to estimate heritabilities for human copy number variations detected by an oligonucleotide-based array with regression analysis, and they reported that most of them were quite high, which were about more than 0.6. This study suggested that almost all copy number polymorphism was consistent with Mendelian inheritance. On the other hand, in this study, we analyzed the inheritance of the copy number of PERV genes. There are a lot of insertions on the pig genome that originated from viral particles, and PERV is one of them. We had expected that the genes on PERV would follow the Mendelian inheritance law, and it was assumed that the heritabilities for the copy number of the genes would be estimated in quite high values, around one. However, we obtained the estimate of the genomic heritabilities of the genes, ranging from 0.267 ± 0.388 to 0.708 ± 0.309 (Table 1), and in the case of the pedigree-based analysis, the heritability estimate for the copy number of pol was 0.594 ± 0.074, suggesting that the estimates in this study were less than one. The results of this study suggested that the heritability of the PERV copy number was affected by other factors such that the PERV genes could be duplicated in the germline.
In the second experiment, we assessed the genetic trends of the copy number of pol to investigate the effectiveness of genetic selection. When the copy number of the pol gene is assumed to be completely inherited (h2 = 1) and the additive standard deviation until the second generation and the standardized selection intensity in the population are 1.29 and 0.90, respectively, the reduction of the copy number in each generation is assumed to be 1.16 according to the breeder's formula. As a result, we anticipate obtaining pigs having zero copy numbers by about the seventh generation and later. However, in this study, although the copy numbers of pol have high heritability (0.594 ± 0.074 in the genomic analysis and 0.400 ± 0.333 in the pedigree analysis), the heritability would be different from one. In addition, the result from the selection test also indicated that the copy number did not completely inherit parents to piglets. These results indicated that we need more generations to obtain the animals with low PERV copy numbers. On the other hand, the not-one heritability estimates in this study imply that some PERVs are always localized at specific regions in the genome in this breeding population. This is because it is widely accepted that transposable elements such as PERVs have evolved to target specific regions where their insertion does not interfere with host cell function and is advantageous for their reproduction (Sultana et al., 2017). Therefore, it would not be very surprising if PERVs are not randomly distributed across the genome. There is a possibility that a few PERVs would be fixed and not be able to segregate during meiosis. However, in this case, it would be impossible to reduce the number of PERV copies to zero by the conventional animal breeding scheme. There are two possible solutions: (1) to introduce some animals with the low PERV copy number from other provinces into the breeding population in this study, and (2) to use the genome editing, which would disrupt all the PERVs regardless of their location (Niu et al., 2017; Yang et al., 2015).
In conclusion, we obtained the high heritability estimates for the PERV copy numbers in the Vietnamese native breeds. This fact indicated that the PERV genes would be segregated on the genome, and most of them are maintained in heterozygotes. However, despite the high heritability estimates for the copy number of the genes, selection for reducing the copy number in the three generations has worked insufficiently. In addition, the result did not explain completely that all the PERV genes are completely segregated in the genome, and if some loci are fixed as homozygosity on the genome, we will not be able to eliminate the genes from this population completely. In this case, alternative approaches, like genome editing, would be required to target all the PERVs.
ACKNOWLEDGMENTS
The authors would like to thank Satoshi Mikawa for the help on the analysis of this study. This research was partially supported by the Science and Technology Research Partnership for Sustainable Development (SATREPS) [JPMJSA1404] from the Japan Science and Technology Agency (JST)/Japan International Cooperation Agency (JICA) and also by Accelerating Social Implementation for SDGs Achievement (aXis) [JPMJAS2006] from JST.
CONFLICT OF INTEREST STATEMENT
We have no conflict of interest to disclose.




