Unveiling the hub genes associated with ochratoxin A-induced hepatotoxicity in broiler chickens
Abstract
Ochratoxin A (OTA) widely exists in raw food and feed materials and can induce liver damage and toxicity. However, the mechanisms of OTA-induced hepatotoxicity were largely unknown. Thus, our study aimed to uncover the vital genes relevant to OTA-induced hepatotoxicity in broiler chickens. Gene expression data of chicken embryo primary hepatocytes (CEPHs) in OTA-treated and control groups were obtained from the GEO database. Totally 1407 differentially expressed genes (DEGs) were selected, of which 850 and 557 genes were up- and downregulated in OTA-treated CEPHs. Gene ontology (GO) enrichment revealed that the DEGs were in connection with various biological processes, such as signal transduction, extracellular matrix organization, axon guidance, cell division, cholesterol homeostasis, proteolysis, microtubule cytoskeleton organization, and chromosome segregation. Pathway enrichment showed that the DEGs were related to metabolic pathways, ferroptosis, calcium, FoxO, Wnt, cell cycle, apoptosis, calcium, and cell adhesion molecules signaling pathways. Furthermore, the hub genes, including CDK1, DLGAP5, KIF2C, VCL, ITGB3, and ZYX, were identified as hub genes potentially contributing to OTA-induced hepatotoxicity. Taken together, this study provides valuable insights into the mechanisms underlying OTA-induced hepatotoxicity in broiler chickens.
1 INTRODUCTION
Ochratoxin A (OTA), widely existing in food and feed materials, induces liver injury and toxicity. Yu et al. (2018) reported that OTA exposure decreased cell viability, accelerated cell apoptosis, and increased the activities of serum aspartate transaminase (AST) and alanine transaminase (ALT) in chicken primary hepatocytes. Pyo et al. (2020) investigated that co-exposure of OTA and acrylamide significantly increased the expressions of phase I reaction-related genes (CYP1A1 and CYP1A2) and apoptosis-associated genes (Bax and CASP3), suggesting that OTA and acrylamide combination contributed to nephrotoxicity and hepatotoxicity. Wang et al. (2020) found that OTA led to apoptosis and increased the expressions of CAP3 and Box in rat liver cells. Xia et al. (2021) revealed that OTA-induced liver inflammation and damage by decreasing inflammatory cell infiltration and increasing the activities of AST, ALT, and alkaline phosphatase in duck serum. OTA exposure increased CASP3 expression while decreased BCL2 expression in chicken primary hepatocytes (Yu et al., 2018). Yang et al. (2023) indicated that OTA exposure increased the levels of AST, TNF-α, IL-6, and IL-1β in mouse serum.
OTA exposure induces oxidative stress in liver tissue. Yu et al. (2018) found that OTA exposure decreased hepatic superoxide dismutase (SOD) and glutathione, increased lipid peroxidation product malondialdehyde in chicken primary hepatocytes. Zhang et al. (2022) found that OTA exposure caused oxidative stress and inflammatory response in rabbit liver, which was characterized by decreasing the activities of SOD, glutathione peroxidase (GSH-Px), and total antioxidant capacity (T-AOC), while increasing the contents of malonaldehyde (MDA), reactive oxygen species, IL-1β, IL-6, and TNF-α. Yang et al. (2023) reported that the activities of T-AOC and SOD were markedly inhibited in the serum and liver of OTA-treated mice. Zhai et al. (2020) reported that the diet with 2 mg/kg OTA inhibited liver catalase activity in 1-day-old Pekin ducklings.
Previous researches have suggested various genes, such as CDK1, DLGAP5, KIF2C, VCL, ITGB3, and ZYX, contributed to liver disease. Are these genes involved in OTA-induced hepatotoxicity? The question remains to be revealed. For instance, Hao et al. (2021) elaborated that CDK1 overexpression caused poor overall survival time for liver cancer patients. In hepatocellular carcinoma (HCC) tissue, DLGAP5 was upregulated and negatively correlated with overall survival time (Huang et al., 2021). KIF2C upregulation promoted cell proliferation and invasion in HCC (Wei et al., 2021). VCL-positive quiescent hepatic stellate cells (HSCs) number was increased in fetal livers (Kawai et al., 2003). ITGB3 gene participated in acute liver failure (Lin et al., 2022). ZYX expression was increased when HCC was recurred after resection (Sy et al., 2006). However, the potential mechanisms of OTA-induced liver toxicity in broiler chickens are unclear. Thus, the present research was to uncover the hub genes leading to hepatotoxicity caused by OTA exposure in broiler chickens.
2 MATERIALS AND METHODS
2.1 Cell isolation, OTA treatment, total RNA extraction, and RNA sequencing
This section was conducted by Sun et al. (2019). In brief, 18-day embryonated specific-pathogen-free Arbor Acres broiler eggs were used to isolate chicken embryo primary hepatocytes (CEPHs). The cells were cultured in a humidified incubator at 37°C with 5.0% CO2. The CEPHs were treated with 5 μg/mL OTA. After 48 h, total RNA was extracted from OTA-treated and untreated CEPHs using Trizol reagent (Invitrogen, Carlsbad, CA, USA). The concentration, purity, and quality of total RNA were assessed. Total RNA was reverse-transcribed into cDNA with oligo d(T) and random primers. The RNA sequencing analysis was then performed on an Illumina HiSeq X Ten platform.
2.2 Identification of differentially expressed genes (DEGs)
Aiming to screen the DEGs in CEPHs, the gene expression analysis was conducted using DESeq R package (1.18.0) based on the data obtained from the GEO database. DEGs were identified based on|log2Fold Change (FC)| > 1 and p < 0.05. Finally, the DEGs were saved for further analysis.
2.3 Gene ontology (GO) and signaling pathway analysis
To gain a better understanding of the DEGs, DAVID database was used to perform GO analysis. KOBAS tool (Version 3.0) was used to conduct KEGG and Reactome analyses to investigate the signaling pathways associated with the DEGs.
2.4 Protein–protein interaction (PPI) network
To explore the interactions among the DEGs, the STRING database was used to construct the PPI networks. The PPI networks were visualized using Cytoscape software (Version 3.8.0).
2.5 Hub genes and their functions
To identify the hub genes, CytoHubba plug-in was used to analyze the PPI networks with Cytoscape software (Version 3.8.0). Finally, their functions were summarized using previous literature, the NCBI database, and GeneCards.
3 RESULTS
3.1 Overview of the genes in CEPHs
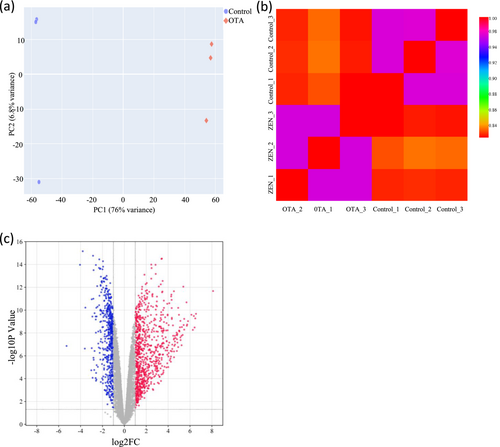
The expression profile of the detected genes in CEPHs treated with and without OTA was shown in Table 1. UMAP and Pearson correlation were shown in Figure 1a,b, respectively. A total of 1407 DEGs were identified with 557 downregulated and 850 upregulated genes in OTA-treated CEPHs, as shown in Data S1 and S2. The volcano plot in Figure 1c represented the genes identified in the CEPHs. Finally, the top 30 up- and downregulated genes in OTA-treated CEPHs were listed in Tables 2 and 3, respectively.
| Total | Downregulated | Upregulated |
|---|---|---|
| 1407 | 557 | 850 |
| Gene symbol | log2FC | p-Value | Description |
|---|---|---|---|
| MYLK2 | 8.11 | 2.13E-12 | Myosin light chain kinase 2 |
| LOC396365 | 6.55 | 2.11E-10 | Preprogastrin |
| TACR2 | 6.48 | 5.33E-10 | Tachykinin receptor 2 |
| CYP2AB1 | 6.42 | 3.30E-09 | Cytochrome P450, family 2, subfamily AB, polypeptide 1 |
| IL12A | 6.35 | 3.28E-10 | Interleukin 12A |
| DFNB59 | 6.26 | 5.39E-09 | Deafness, autosomal recessive 59 |
| LVRN | 6.13 | 2.53E-10 | Laeverin |
| LMOD3 | 5.82 | 9.26E-10 | Leiomodin 3 |
| CYP2AB2 | 5.74 | 1.83E-09 | Cytochrome P450, family 2, subfamily AB, polypeptide 2 |
| TBX4 | 5.71 | 2.31E-09 | T-box 4 |
| TLR7 | 5.68 | 2.08E-11 | Toll like receptor 7 |
| BEST1 | 5.47 | 5.86E-10 | Bestrophin 1 |
| GGT2 | 5.47 | 5.09E-09 | Gamma-glutamyltransferase 2 |
| NR5A1 | 5.42 | 2.02E-09 | Nuclear receptor subfamily 5 group A member 1 |
| RAB44 | 5.39 | 8.79E-13 | RAB44, member RAS oncogene family |
| HAS3 | 5.33 | 1.49E-08 | Hyaluronan synthase 3 |
| CRYBB3 | 5.26 | 3.04E-09 | Crystallin beta B3 |
| TACSTD2 | 5.23 | 1.19E-07 | Tumor associated calcium signal transducer 2 |
| LMX1A | 5.22 | 1.80E-08 | LIM homeobox transcription factor 1, alpha |
| CHRNA3 | 5.14 | 4.21E-08 | Cholinergic receptor nicotinic alpha 3 subunit |
| CELF3 | 5.12 | 8.78E-10 | CUGBP Elav-like family member 3 |
| OTX1 | 5.12 | 2.45E-08 | Orthodenticle homeobox 1 |
| SLCO1B1 | 5.08 | 3.20E-09 | Solute carrier organic anion transporter family member 1B1 |
| KLHL35 | 5.03 | 7.41E-09 | Kelch like family member 35 |
| SCARA5 | 5.01 | 6.24E-09 | Scavenger receptor class A member 5 |
| MESP2 | 5.01 | 6.37E-09 | Mesoderm posterior bHLH transcription factor 2 |
| KLHL33 | 4.99 | 2.57E-10 | Kelch like family member 33 |
| PI16 | 4.92 | 6.33E-11 | Peptidase inhibitor 16 |
| LOC112530787 | 4.92 | 1.73E-07 | Small Cajal body-specific RNA 8 |
| NEIL2 | 4.85 | 7.02E-08 | Nei like DNA glycosylase 2 |
| Gene symbol | log2FC | p-Value | Description |
|---|---|---|---|
| FSHB | −5.28 | 1.38E-07 | Follicle stimulating hormone beta subunit |
| HSPB7 | −4.05 | 1.07E-14 | Heat shock protein family B (small) member 7 |
| KPNA7 | −3.8 | 6.95E-16 | Karyopherin subunit alpha 7 |
| WNT11B | −3.61 | 2.35E-07 | Wingless-type MMTV integration site family, member 11b |
| CD3E | −3.56 | 6.04E-11 | CD3e molecule |
| CPA5 | −3.02 | 1.86E-05 | Carboxypeptidase A5 |
| ITGB4 | −2.98 | 1.13E-11 | Integrin subunit beta 4 |
| GPR4 | −2.98 | 2.13E-07 | G-protein-coupled receptor 4 |
| SLC6A15 | −2.92 | 3.67E-10 | Solute carrier family 6 member 15 |
| LOC418667 | −2.9 | 8.42E-05 | Granulocyte-macrophage colony-stimulating factor receptor subunit alpha-like |
| TSPAN2 | −2.86 | 1.73E-15 | Tetraspanin 2 |
| FSCN1 | −2.83 | 1.00E-11 | Fascin actin-bundling protein 1 |
| SYNPO2L | −2.82 | 1.48E-06 | Synaptopodin 2 like |
| CCDC80 | −2.81 | 3.16E-14 | Coiled-coil domain containing 80 |
| HTR2B | −2.8 | 2.22E-06 | 5-hydroxytryptamine receptor 2B |
| RAB39A | −2.75 | 1.74E-05 | RAB39A, member RAS oncogene family |
| APOBEC2 | −2.72 | 1.85E-10 | Apolipoprotein B mRNA editing enzyme catalytic subunit 2 |
| CLEC5A | −2.72 | 3.78E-07 | C-type lectin domain family 5, member A |
| SLC22A3 | −2.62 | 3.63E-10 | Solute carrier family 22 member 3 |
| BDKRB1 | −2.62 | 8.84E-05 | Bradykinin receptor B1 |
| BCAS1 | −2.61 | 6.56E-10 | Breast carcinoma amplified sequence 1 |
| RAI14 | −2.6 | 1.11E-14 | Retinoic acid induced 14 |
| LOC427933 | −2.6 | 9.44E-09 | Sulfotransferase 6B1-like |
| MIR1661 | −2.59 | 1.53E-04 | MicroRNA 1661 |
| AHNAK2 | −2.58 | 3.17E-05 | AHNAK nucleoprotein 2 |
| CCL17 | −2.56 | 6.23E-11 | C-C motif chemokine ligand 17 |
| HAPLN2 | −2.55 | 1.21E-04 | Hyaluronan and proteoglycan link protein 2 |
| LAMA1 | −2.53 | 3.59E-10 | Laminin subunit alpha 1 |
| ETV7 | −2.53 | 6.58E-06 | ETS variant 7 |
| FAP | −2.48 | 7.90E-13 | Fibroblast activation protein alpha |
3.2 GO enrichment of the DEGs
The upregulated DEGs in OTA-treated CEPHs appeared to be involved in signal transduction, extracellular matrix organization, axon guidance, cell division, cholesterol homeostasis, proteolysis, microtubule cytoskeleton organization, chromosome segregation, osteoblast differentiation, muscle contraction, cell–cell signaling, xenobiotic metabolic process, cytolysis, bicellular tight junction assembly, lipid transport, blood coagulation, sodium ion transmembrane transport, cell fate specification, mitotic spindle organization, mitotic chromosome condensation, triglyceride homeostasis, monocarboxylic acid transport, proteoglycan biosynthetic process, cholesterol efflux, and cell maturation (Figure 2a and Data S3).
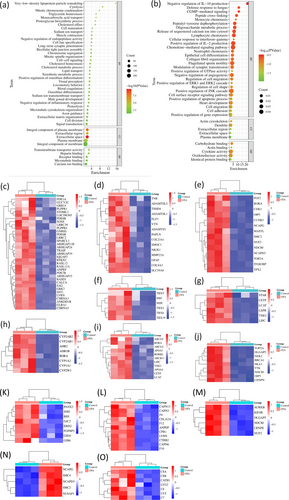
Furthermore, the downregulated DEGs in OTA-administrated CEPHs were perhaps linked to cell adhesion, JNK cascade, monocyte chemotaxis, neutrophil chemotaxis, epithelial cell differentiation, collagen fibril organization, cell migration, cGMP-mediated signaling, peptidyl-tyrosine dephosphorylation, oligosaccharide metabolic process, lymphocyte chemotaxis, synaptic transmission modulation; GTPase activity, apoptotic process, IL-2 production; regulation of cell shape, and migration (Figure 2b and Data S4). Figure 2c–o showed the heatmaps to reveal the signaling pathways about the DEGs between the two groups, including signal transduction, extracellular matrix organization, cell division, cell fate specification, cholesterol metabolic process, xenobiotic metabolic process, cholesterol homeostasis, chromosome segregation, cell–cell signaling, proteolysis, mitotic spindle organization, mitotic chromosome condensation, and cytolysis signaling pathways, respectively.
3.3 KEGG enrichment of the DEGs
In OTA-treated CEPHs, the upregulated genes were potentially associated with various signaling pathways, such as metabolic pathways, ferroptosis, calcium, FoxO, Wnt, adipocytokine, ECM-receptor interaction, tight junction, steroid hormone biosynthesis, cell cycle, ABC transporters, actin cytoskeleton regulation, and the metabolisms of retinol, vitamin B6, tryptophan, nicotinate, nicotinamide, arachidonic acid, glycerophospholipid, taurine, hypotaurine, glycine, serine, threonine, histidine, purine, and drug (Figure 3a and Data S5).
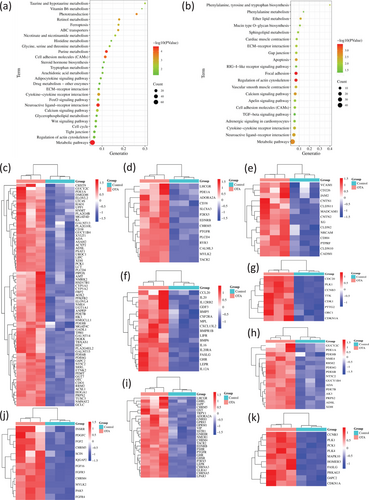
Figure 3b and Data S6 indicated that in OTA-administrated CEPHs the downregulated genes might be closely related to various signaling pathways, such as metabolic pathways, apoptosis, calcium, Gap junction, apelin, cell adhesion molecules, neuroactive ligand-receptor interaction, focal adhesion, RIG-I-like receptor, TGF-β, actin cytoskeleton regulation, ECM-receptor interaction, and cytokine-cytokine receptor interaction; the metabolisms of ether lipid, sphingolipid, and phenylalanine; the biosynthesis of mucin type O-glycan, phenylalanine tyrosine, and tryptophan.
Moreover, Figure 3c–k showed the heatmaps depicting the upregulated genes in OTA-treated CEPHs in metabolic pathways, calcium, cell adhesion molecules, cytokine-cytokine receptor interaction, cell cycle, purine metabolism, neuroactive ligand-receptor interaction, actin cytoskeleton regulation, and FoxO signaling pathways, respectively.
3.4 Reactome enrichment of the DEGs
In OTA-treated CEPHs, the upregulated genes might participate in many signaling pathways, including immune system, small molecules transport, GPCR downstream signaling, innate immune system, metabolism, signal transduction, hemostasis, cell cycle, signaling by GPCR, vesicle-mediated transport, signaling by Rho GTPases, G alpha (i) signaling events, SLC-mediated transmembrane transport, M Phase, membrane trafficking, mitotic prometaphase, sister chromatid cohesion resolution, mitotic anaphase, cell cycle checkpoints, sister chromatids separation, mitotic metaphase and anaphase, RHO GTPase effectors, mitotic spindle checkpoint, signaling by interleukins, amino acids and derivative metabolism, signaling by Wnt, G alpha (s) signaling events, MAPK1/MAPK3 signaling, FLT3 signaling, and MAPK family signaling cascades (Figure 4a).
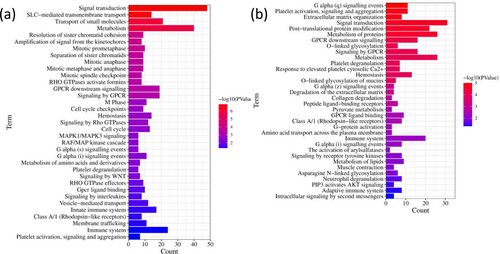
In addition, in OTA-incubated CEPHs, the downregulated genes might be closely related to signal transduction, metabolism, protein metabolism, immune system, hemostasis, signaling by GPCR, GPCR downstream signaling, neutrophil degranulation, adaptive immune system, collagen degradation, pyruvate metabolism, arylsulfatases activation, G-protein activation, opioid signaling, protein folding, sphingolipid metabolism, G alpha (12/13) signaling events, chaperonin-mediated protein folding, extranuclear estrogen signaling, G alpha (z) signaling events, extracellular matrix degradation, platelet degranulation, peptide ligand-binding receptors, asparagine N-linked glycosylation, and mucins O-linked glycosylation (Figure 4b).
3.5 Protein classification for the DEGs
In OTA-treated CEPHs, the upregulated DEGs played a crucial role in hydrolase, transferase, protease, transporter, lyase, cytoskeletal protein, ion channel, kinase, phosphodiesterase, cyclase, metalloprotease, protease inhibitor, phospholipase, oxygenase, p53-like transcription factor, guanylate cyclase, peptide hormone, complement component, actin-binding motor protein, nucleotide kinase, ATP-binding cassette, secondary carrier transporter, microtubule binding motor protein, protein modifying enzyme, scaffold/adaptor protein, transmembrane signal receptor, and metabolite interconversion enzyme (Figure 5a).
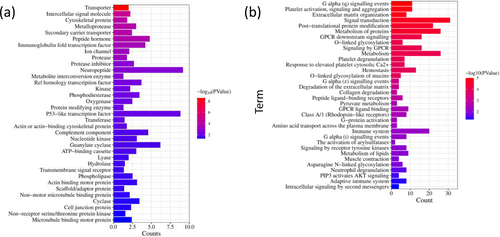
Additionally, in OTA-administrated CEPHs, the downregulated DEGs might be associated with protein modifying enzyme, intercellular signal molecule, cytoskeletal protein, intermediate filament, metabolite interconversion enzyme, extracellular matrix glycoprotein, isomerase, extracellular matrix protein, guanylate cyclase, neurotrophic factor, and actin or actin-binding cytoskeletal protein (Figure 5b).
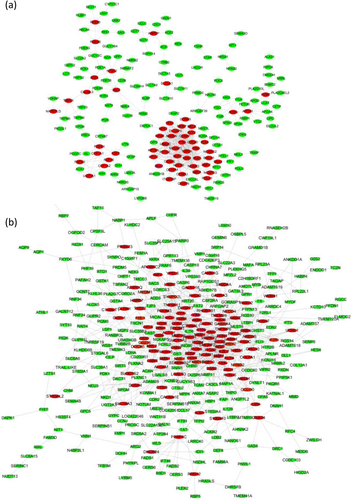
3.6 PPI network
To further selected the hub genes closely associated with OTA-induced hepatotoxicity, we constructed the PPI networks for the DEGs. The upregulated genes, such as CDK1, DLGAP5, KIF2C, TTK, NUSAP1, AURKA, NUF2, CENPF, PBK, MELK, and CDC20, likely played an essential role in hepatotoxicity induced by OTA (Figure 6a). Additionally, the downregulated genes, including VCL, ITGB3, ZYX, RHOB, PDLIM3, ACTC1, FN1, FYN, SYNPO2L, TEK, NGF, and BDNF, might participate in OTA-induced hepatotoxicity (Figure 6b).
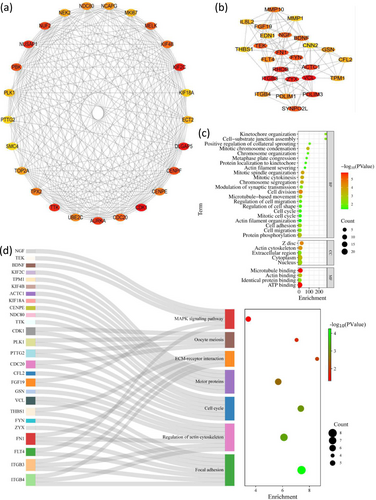
3.7 Hub genes and their functions
For the purpose of selecting the hub genes, CytoHubba plug-in was used to analyze PPI network. In OTA-treated CEPHs, the top 25 upregulated hub genes included CDK1, DLGAP5, and KIF2C, et al (Figure 7a and Table 4). Additionally, the top 25 downregulated hub genes in OTA-administrated CEPHs included VCL, ITGB3, and ZYX, et al (Figure 7b and Table 5). Furthermore, GO enrichment analysis suggested that the 50 hub genes might be associated with cell division, protein phosphorylation, mitotic spindle organization, mitotic cytokinesis, cell adhesion, chromosome segregation, mitotic chromosome condensation, microtubule-based movement, cell-substrate junction assembly, kinetochore organization, synaptic transmission modulation, cell migration, cell migration regulation, chromosome organization, cell shape regulation, protein localization to kinetochore, actin-filament severing, cell cycle, and positive regulation of collateral sprouting (Figure 7c). Moreover, KEGG enrichment analysis suggested that the 50 hub genes might be associated with MAPK, focal adhesion, actin cytoskeleton regulation, cell cycle, motor proteins, and ECM-receptor interaction signaling pathways (Figure 7d).
| Gene symbol | Full name | Functions |
|---|---|---|
| CDK1 | Cyclin-dependent kinase 1 | CDK1 plays a key role in the control of the eukaryotic cell cycle by modulating the centrosome cycle as well as mitotic onset; promotes G2-M transition, regulates G1 progress and G1-S transition via association with multiple interphase cyclins. |
| DLGAP5 | DLG associated protein 5 | DLGAP5 is predicted to enable microtubule binding activity and involved in several processes, including centrosome localization; kinetochore assembly; and mitotic spindle organization. |
| KIF2C | Kinesin family member 2C | KIF2C encodes a kinesin-like protein that functions as a microtubule-dependent molecular motor. The encoded protein can depolymerize microtubules at the plus end, thereby promoting mitotic chromosome segregation. |
| TTK | TTK protein kinase | TTK encodes a dual specificity protein kinase with the ability to phosphorylate tyrosine, serine and threonine. Associated with cell proliferation, this protein is essential for chromosome alignment at the centromere during mitosis and is required for centrosome duplication. |
| NUSAP1 | Nucleolar and spindle-associated protein 1 | NUSAP1 is a nucleolar-spindle-associated protein that plays a role in spindle microtubule organization, RNA binding, and microtubule binding. |
| AURKA | Aurora kinase A | The protein encoded by AURKA is a cell cycle-regulated kinase that appears to be involved in microtubule formation and/or stabilization at the spindle pole during chromosome segregation. The protein is found at the centrosome in interphase cells and at the spindle poles in mitosis. |
| NUF2 | Kinetochore protein Nuf2 | NUF2 acts as a component of the essential kinetochore-associated NDC80 complex, which is required for chromosome segregation and spindle checkpoint activity. |
| CENPF | Centromere protein F | CENPF encodes a protein that associates with the centromere-kinetochore complex. The protein is a component of the nuclear matrix during the G2 phase of interphase. In late G2, the protein associates with the kinetochore and maintains this association through early anaphase. |
| PBK | PDZ binding kinase | PBK encodes a serine/threonine-protein kinase related to the dual specific mitogen-activated protein kinase kinase (MAPKK) family. Evidence suggests that mitotic phosphorylation is required for its catalytic activity. |
| MELK | Maternal embryonic leucine zipper kinase | MELK is involved in apoptotic process, cell population proliferation, and protein autophosphorylation. |
| CDC20 | Cell division cycle 20 | CDC20 protein appears to act as a regulatory protein interacting with several other proteins at multiple points in the cell cycle. It is required for two microtubule-dependent processes, nuclear movement prior to anaphase, and chromosome separation. |
| KIF4B | Kinesin family member 4B | KIF4B is an intronless retrocopy of kinesin family member 4A. The protein encoded by this gene is a microtubule-based motor protein that plays vital roles in anaphase spindle dynamics and cytokinesis. |
| CENPE | Centromere protein E | CENPE is a kinesin-like motor protein that accumulates in the G2 phase of the cell cycle. |
| TPX2 | TPX2 microtubule nucleation factor | TPX2 enables importin-alpha family protein binding activity and protein kinase binding activity. It is involved in protein kinase activity activation, microtubule cytoskeleton organization, and negative regulation of microtubule depolymerization. |
| UBE2C | Ubiquitin-conjugating enzyme E2 C | The modification of UBE2C protein with ubiquitin is an important cellular mechanism for targeting abnormal or short-lived proteins for degradation. UBE2C involves at least three classes of enzymes: Ubiquitin-activating enzymes, ubiquitin-conjugating enzymes, and ubiquitin-protein ligases. |
| NDC80 | Kinetochore protein NDC80 homolog | NDC80 acts as a component of the essential kinetochore-associated NDC80 complex, which is required for chromosome segregation and spindle checkpoint activity. It is required for kinetochore integrity and the organization of stable microtubule binding sites in the outer plate of the kinetochore. |
| NCAPG | Non-SMC condensin I complex subunit G | This gene encodes a subunit of the condensin complex, which is responsible for the condensation and stabilization of chromosomes during mitosis and meiosis. Phosphorylation of the encoded protein activates the condensin complex. There are pseudogenes for this gene on chromosomes 8 and 15. Alternative splicing results in multiple transcript variants. |
| TOP2A | DNA topoisomerase 2-alpha | Control of topological states of DNA by transient breakage and subsequent rejoining of DNA strands. Topoisomerase II makes double- strand breaks. Essential during mitosis and meiosis for proper segregation of daughter chromosomes. May play a role in the regulation of circadian rhythm. |
| NEK2 | NIMA related kinase 2 | This gene encodes a serine/threonine-protein kinase that is involved in mitotic regulation. This protein is localized to the centrosome and undetectable during G1 phase but accumulates progressively throughout the S phase, reaching maximal levels in late G2 phase. |
| ECT2 | Epithelial cell transforming 2 | ECT2 protein is a guanine nucleotide exchange factor and transforming protein that is related to Rho-specific exchange factors and yeast cell cycle regulators. The expression of this gene is elevated with the onset of DNA synthesis and remains elevated during G2 and M phases. |
| MKI67 | Marker of proliferation Ki-67 | MKI67 is involved in regulation of chromosome segregation and regulation of mitotic nuclear division. |
| PLK1 | Polo like kinase 1 | Depletion of PLK1 protein in cancer cells dramatically inhibited cell proliferation and induced apoptosis; hence, it is a target for cancer therapy. |
| KIF18A | Kinesin family member 18A | KIF18A is a member of the kinesin superfamily of microtubule-associated molecular motors. |
| PTTG2 | Pituitary tumor-transforming gene 2 protein | Predicted to enable SH3 domain binding activity. Predicted to be involved in homologous chromosome segregation and negative regulation of mitotic sister chromatid separation. Predicted to be located in cytoplasm. Predicted to be active in nucleus (provided by Alliance of Genome). |
| SMC4 | Structural maintenance of chromosomes 4 | SMC4 plays a role in two changes in chromosome structure during mitotic segregation of chromosomes- chromosome condensation and sister chromatid cohesion. The protein encoded by SMC4 is likely a subunit of the 13S condensin complex, which is involved in chromosome condensation. |
| Gene symbol | Full name | Functions |
|---|---|---|
| VCL | Vinculin | VCL is a cytoskeletal protein associated with cell–cell and cell-matrix junctions, where it is thought to function as one of several interacting proteins involved in anchoring F-actin to the membrane. |
| ITGB3 | Integrin subunit beta 3 | The ITGB3 protein product is the integrin beta chain beta 3. Integrins are integral cell-surface proteins composed of an alpha chain and a beta chain. ITGB3 is known to participate in cell adhesion. |
| ZYX | Zyxin | ZYX may function as a messenger in the signal transduction pathway that mediates adhesion-stimulated changes in gene expression and may modulate the cytoskeletal organization of actin bundles. |
| RHOB | Ras homolog family member B | RHOB is predicted to enable GTP binding activity; GTPase activity; and protein kinase binding activity. It is involved in several processes, including cellular response to hydrogen peroxide; cellular response to ionizing radiation; and regulation of cell migration. |
| PDLIM3 | PDZ and LIM domain 3 | The protein encoded by PDLIM3 contains a PDZ domain and a LIM domain, indicating that it may be involved in cytoskeletal assembly. The protein has been shown to bind the spectrin-like repeats of alpha-actinin-2 and to colocalize with alpha-actinin-2 at the Z lines of skeletal muscle. |
| ACTC1 | Actin alpha cardiac muscle 1 | ACTC1 is highly conserved proteins that are involved in various types of cell motility. Polymerization of globular actin (G-actin) leads to a structural filament (F-actin) in the form of a two-stranded helix. The protein encoded by this gene belongs to the actin family which is comprised of three main groups of actin isoforms, alpha, beta, and gamma. |
| FN1 | Fibronectin 1 | FN1 encodes fibronectin, a glycoprotein present in a soluble dimeric form in plasma and in a dimeric or multimeric form at the cell surface and in extracellular matrix. FN1 is involved in cell adhesion and migration processes including embryogenesis, wound healing, and blood coagulation. |
| FYN | FYN proto-oncogene, src family tyrosine kinase | FYN is a member of the protein-tyrosine kinase oncogene family. It encodes a membrane-associated tyrosine kinase that has been implicated in the control of cell growth. |
| SYNPO2L | Synaptopodin 2 like | Predicted to enable actin-binding activity. Predicted to be involved in several processes, including positive regulation of Rho protein signal transduction; positive regulation of stress fiber assembly; and sarcomere organization. |
| TEK | TEK receptor tyrosine kinase | TEK encodes a receptor that belongs to the protein-tyrosine kinase Tie2 family. The encoded protein possesses a unique extracellular region that contains two immunoglobulin-like domains, three epidermal growth factor (EGF)-like domains and three fibronectin type III repeats. The ligand angiopoietin-1 binds to this receptor and mediates a signaling pathway that functions in embryonic vascular development. |
| NGF | Nerve growth factor | NGF is a member of the NGF-beta family and encodes a secreted protein which homodimerizes and is incorporated into a larger complex. This protein has the complex is involved in the regulation of growth and the differentiation of sympathetic and certain sensory neurons. |
| BDNF | Brain derived neurotrophic factor | BDNF encodes a member of the nerve growth factor family of proteins. |
| MMP10 | Matrix metallopeptidase 10 | MMP10 is involved in the breakdown of extracellular matrix in normal physiological processes, such as embryonic development, reproduction, and tissue remodeling, as well as in disease processes, such as arthritis and metastasis. |
| FGF19 | Fibroblast growth factor 19 | FGF19 is a member of the fibroblast growth factor (FGF) family. FGF family members are involved in a variety of biological processes including embryonic development cell growth, morphogenesis, and tissue repair. |
| FLT4 | Fms related receptor tyrosine kinase 4 | FLT4 encodes a tyrosine kinase receptor for vascular endothelial growth factors C and D. The protein is thought to be involved in lymphangiogenesis and maintenance of the lymphatic endothelium. |
| PDLIM1 | PDZ and LIM domain 1 | PDLIM1 is a cytoplasmic protein associated with the cytoskeleton. The protein may function as an adapter to bring other LIM-interacting proteins to the cytoskeleton. |
| ITGB4 | Integrin subunit beta 4 | Integrins mediate cell-matrix or cell–cell adhesion and transduced signals that regulate gene expression and cell growth. ITGB4 encodes the integrin beta 4 subunit, a receptor for the laminins. ITGB4 is likely to play a pivotal role in the biology of invasive carcinoma. |
| GSN | Gelsolin | The protein encoded by GSN binds to the “plus” ends of actin monomers and filaments to prevent monomer exchange. The encoded calcium-regulated protein functions in both assembly and disassembly of actin filaments. |
| TPM1 | Tropomyosin 1 | TPM1 is a member of the tropomyosin family of highly conserved, widely distributed actin-binding proteins involved in the contractile system of striated and smooth muscles and the cytoskeleton of nonmuscle cells. It also functions in association with the troponin complex to regulate the calcium-dependent interaction of actin and myosin during muscle contraction. |
| CFL2 | Cofilin 2 | CFL2 encodes an intracellular protein that is involved in the regulation of actin-filament dynamics. This protein is a major component of intranuclear and cytoplasmic actin rods. It controls actin polymerization and depolymerization in a pH-dependent manner. |
| IL8L2 | Interleukin 8-like 2 | - |
| EDN1 | Endothelin 1 | EDN1 encodes a preproprotein that is proteolytically processed to generate a secreted peptide that belongs to the endothelin/sarafotoxin family. |
| MMP1 | Matrix metallopeptidase 1 | MMP1 protein is involved in the breakdown of extracellular matrix in normal physiological processes, such as embryonic development, reproduction, and tissue remodeling, as well as in disease processes. |
| THBS1 | Synaptopodin 2 like | THBS1 was predicted to enable actin-binding activity and involved in several processes, including positive regulation of Rho protein signal transduction and positive regulation of stress fiber assembly. |
| CNN2 | Calponin 2 | The protein encoded by CNN2, which can bind actin, calmodulin, troponin C, and tropomyosin, may function in the structural organization of actin filaments. The encoded protein could play a role in smooth muscle contraction and cell adhesion. |
4 DISCUSSION
OTA exists ubiquitously in raw food and feed materials, resulting in liver damage and toxicity. However, up to now, the studies investigating the genes related to OTA-induced hepatotoxicity using transcriptome techniques remain limited. Therefore, this study was aimed to uncover the vital genes linked to the hepatotoxicity induced by OTA exposure in broiler. Our research suggested various genes, such as CDK1, DLGAP5, KIF2C, VCL, ITGB3, and ZYX, were contribute to OTA-induced hepatotoxicity in broiler chickens.
CDK1 (cyclin-dependent kinase 1) might be associated with OTA-induced hepatotoxicity. CDK1 overexpression was linked to poor overall survival time for the liver cancer patients (Hao et al., 2021). Lv et al. (2021) found that CDK1 phosphorylation mediated liver injury induced by lipopolysaccharide/D-galactosamine. In this study, we found that CDK1 gene was a gene associated with cell cycle pathway (Data S5) and cell division biological process (Data S3), and OTA exposure significantly increased its expression in CEPHs (Figure 2e), which was coincided with the previous reports. Qi et al. (2018) found that OTA exposure induced apoptosis of rat renal tubular epithelial cell (NRK-52E cell) by increasing the expression of CAP3 and CDK1 genes. Similarly, in the study from Adler et al. (2009), they found 70 and 210 μg/kg OTA administration increased the expressions of CDK1, mitotic protein kinases (PLK-1 and AURKB), and several cyclins and cyclin-dependent kinase inhibitors (including TOP2) in rat kidneys.
DLGAP5, named DLG associated protein 5, participated in liver cell health and disease. Liao et al. (2013) revealed that compared to normal liver tissue, the expression of DLGAP5 gene was increased in HCC. Also, DLGAP5 silencing by RNA interference remarkably inhibited growth and migration of HCC cells in vitro. DLGAP5 was one of 10 hub genes associated with liver cancer (Hao et al., 2021). Huang et al. (2021) identified 14 hub genes (including DLGAP5) in HCC, whose expression was upregulated and inversely correlated with overall survival time and immune infiltration cells including monocytes. However, it was positively correlated with regulatory T follicular helper cells, T cells, and macrophages M0. In the present study, we observed that DLGAP5 participated in mitotic spindle organization and chromosome segregation biological processes (Data S3), and its expression in CEPHs was remarkedly increased after OTA treatment, which was consistent with previous reports from Matsuoka et al. (2002) and White et al. (2005).
KIF2C, alias kinesin family member 2C, participates in liver cell proliferation, migration, apoptosis, and invasion as well as liver disease occurrence. KIF2C overexpression promoted HCC cell proliferation, migration, invasion, and metastasis by binding TBC1D7 gene (named TBC1 domain family member 7), disrupting TSC complex formation, and enhancing mTORC1 signal transduction (Wei et al., 2021). KIF2C upregulation was linked to the poor overall survival, stage, pathological grade, and disease-free survival of HCC patients (Ji et al., 2020; Li et al., 2020). In our research, we found KIF2C was associated with microtubule binding (Data S3) and OTA treatment significantly increased KIF2C expression in CEPHs, which was in line with the findings from Moon et al. (2021) that KIF2C participated in microtubule dynamics, which was important for cell motility and migration by regulating endocytosis and transport of focal adhesion components. Thus, we speculated KIF2C might participate in OTA-induced hepatotoxicity by regulating microtubule dynamics of CEPHs.
VCL (vinculin) is a cytoskeletal protein and associated with liver injury. In the chicken liver nuclei, VCL protein might interacted with histone and actin specific proteases (Panda et al., 2021). The quiescent HSCs would be activated when the liver was impaired. Subsequently, intense immunoreaction of vinculin protein was observed in activated HSCs (Kawai et al., 2003). In addition, the number of VCL-positive quiescent HSCs was gradually increased in fetal livers during gestation (Kawai et al., 2003). In this study, we found VCL was associated with cell adhesion, actin cytoskeleton, and cell migration regulation (Data S4), which was in agreement with the study from Panda et al. (2021). In addition, a decrease in VCL expression in CEPHs after OTA treatment might reveal that the cells were in the repair phase after injury.
ITGB3 (integrin subunit beta 3) might be participate in OTA-induced hepatotoxicity. In this study, ITGB3 was found to be linked to the biological processes (including cell migration) and multiple signaling pathways (such as signal transduction and integrin cell-surface interactions), as shown in Data S4 and S6. These findings were consistent with previous reports. Lin et al. (2022) found that VWF/RAP1B/ITGB3 signaling mediated the process of ameliorating acute liver failure. Lee et al. (2022) also reported that ITGB3 regulated PPAR alpha signaling and extracellular matrix accumulation in activated HSCs in multicellular hepatic spheroids. Also, it was associated with liver fibrosis and hepatic lipid accumulation in nonalcoholic steatohepatitis. ITGB3 knockdown decreased oil accumulation and oxidative stress levels and reversed the downregulation of apoptosis by inhibiting PIK3R1/Akt pathway activation (Li et al., 2022).
ZYX (zyxin) is associated with liver health and disease. Sy et al. (2006) indicated that the expression of ZYX gene was increased in HCC tissues. Moreover, in the initial resected tumor, ZYX overexpression was displayed when HCC was recurred after resection. Cai et al. (2023) also found that ZYX expression was increased in HCC tissues. Moreover, ZYX overexpression accelerated cell migration, proliferation, and invasion, while ZYX knockdown had the reverse effects in hepatoma cell lines. In addition, ZYX expression level altered the levels of the proteins linked to cell migration, cycle, and invasion. In this study, we observed that ZYX participated in two biological processes: cellular response to interferon-γ and positive regulation of gene expression, which accorded with early reports from Horras et al. (2011) and Shen et al. (2021). Interferon-γ might modulate hepatocyte fate during liver transplantation, regeneration, hepatitis, fibrosis, and HCC (Horras et al., 2011). Shen et al. (2021) found that interferon-γ induced liver inflammation and damage by activating STAT1 signaling.
Nevertheless, this study existed two disadvantages limiting the significance. Firstly, this research was not sufficient in verifying the results from transcriptomic data. Fortunately, previous studies have validated the results of gene expression in our study. For instance, we found that the expression of AHR2 gene was increased by 3.19 times in OTA-treated liver cells (Data S1), which was verified by Kövesi et al. (2024). Secondly, in the future, special mechanism research for the hub genes is needed to be carried out.
In conclusions, we found that many genes, such as CDK1, DLGAP5, KIF2C, VCL, ITGB3, and ZYX, might contribute to OTA-induced hepatotoxicity. Altogether, this research enhanced our comprehensive understanding about the molecule mechanisms of OTA-induced hepatotoxicity.
ACKNOWLEDGMENTS
This study was supported by The Open Project of State Key Laboratory of Animal Biotech Breeding (No. SKLABB202308), The Open Project of State Key Laboratory of Tea Plant Biology and Utilization (No. SKLTOF20230121), Anhui Provincial Key Research and Development Project (No. 202204c06020076), The University Synergy Innovation Program of Anhui Province (No. GXXT-2023-105), The Open Project of Longyan University & Fujian Provincial Key Laboratory for Prevention and Control of Animal Infectious Diseases and Biotechnology (No. ZDSYS2023003), and The Open Project of Anhui Province Key Laboratory of Embryo Development and Reproductive Regulation (No. FSKFKT011).
CONFLICT OF INTEREST STATEMENT
All authors declare that this research was implemented without any interest conflicts.




