Genetic polymorphisms of calpain1 and calpain3 genes and their effects on growth, carcass, and meat quality traits in Betong chicken (KU line)
Abstract
Betong chicken (KU line) is a slow-growing Thai native chicken used for meat production. The objectives of this study were to identify polymorphisms of the calpain1 (CAPN1) and calpain3 (CAPN3) genes and to investigate their effects on growth, carcass, and meat quality traits in Betong chickens (KU line). A sample of 252 Betong chickens (KU line) was screened for CAPN1 and CAPN3 polymorphisms. The polymorphisms of CAPN1 were detected using gel electrophoresis and DNA sequencing, whereas the polymorphisms of CAPN3 were identified using restriction fragment length polymorphism. Polymorphisms were detected in both CAPN1 (AA, AB, and BB genotypes) and CAPN3 (CC, CT, and TT genotypes). The frequency of the B allele was higher than for the A allele (0.78 and 0.22, respectively) in CAPN1, while the C allelic frequency was higher than for the T allele (0.54 and 0.46, respectively) in CAPN3. The CAPN1 genotype and the combination of the CAPN1 and CAPN3 genotypes could be used as genetic markers for meat lightness. The CAPN3 could be useful for increasing body weight, live weight, and breast meat weight in Betong chickens (KU line).
1 INTRODUCTION
Calpains are Ca2+-dependent cysteine proteases that promote growth and meat quality traits (Felício et al., 2013; Rasouli et al., 2013). Calpains have been classified into two types based on their expression patterns in different tissues: ubiquitous and tissue specific (Sorimachi et al., 1994). The calpain1 gene (CAPN1) was found to be ubiquitously expressed, while the calpain3 gene (CAPN3) was found to be expressed in skeletal muscles (Suzuki et al., 2004). In chicken, the calpain1 plays an important role in muscle growth regulation and postmortem meat proteolysis, whereas the calpain3 is a skeletal muscle-specific protease and regulates myogenesis (Sorimachi & Suzuki, 2001; Zhang et al., 2008). Genetic polymorphisms of CAPN1 were found in Southeast Asian native chickens (Okumura et al., 2006). Zhang et al. (2009) found that haplotypes with the three single-nucleotide polymorphisms (SNPs) (2546C > T, 3535G > A, and 7198C > A) were associated with live weight, carcass weight (CW), breast meat weight (BMW), leg weight (LEG), eviscerated percentage, and breast muscle fiber density using polymerase chain reaction (PCR)-single strand conformation polymorphism. As for CAPN3, the SNPs in this gene were found to be associated with body weight (BW) and carcass traits in five commercial pure lines and four native chicken breeds from the Guangdong and Sichuan provinces in China (Zhang et al., 2009). Therefore, CAPN1 and CAPN3 may be major genes that could be suitable markers for growth, carcass, and meat quality traits in chicken.
Betong chicken (KU line) was developed from a native chicken (Betong) type in southern Thailand. Its meat is especially suitable for use in traditional Thai dishes such as steamed rice topped with boiled chicken, chicken spicy soup with Thai herbs. In Southeast Asia, the selling price of native chicken meat is three times of that of broiler meat and native chicken meat is more favorable in niche markets (Bungsrisawat et al., 2018). Thus, high production of birds with desirable growth, carcass, and meat quality traits results in a high profit for producers. Genetic information on the CAPN1 and CAPN3 may help to improve the growth, carcass, and meat quality traits of Betong chickens. Thus, the aims of this study were to determine the allele and genotype frequencies as well as the polymorphisms of the CAPN1 and CAPN3 and to analyze the effects of their polymorphisms on carcass and meat quality traits in Betong chicken (KU line).
2 MATERIALS AND METHODS
2.1 Animal care
Animal care in this research was reviewed and approved by the Kasetsart University's Institutional Animal Care and Use Committee, Bangkok, Thailand (ACKU60-AGR-003).
2.2 Animal and phenotypic data
Initially, the Betong chicken (KU line) population was established from a poultry farm in southern Thailand. Cocks and hens were selected based on the original breed characteristics, namely, feathering rate, golden brown feather color, and growth performance. Additionally, mating based on full-sib and half-sib relationships was avoided in this closed flock (Sopannarath & Bunchasak, 2015).
In the current study, phenotypic measurements (growth, carcass, and meat quality traits) were tested in two progeny flocks. The progeny flocks were produced by artificially inseminating hens from the same cock (using 27 cocks and 96 hens) on a weekly basis at the poultry farm located at Kasetsart University, Bangkok, Thailand. Identification of pedigree information was conducted by daily collection of the eggs and marking them according to dam number. Prior to incubation, the collected eggs were disinfected with formaldehyde and subsequently placed in an incubator for 21 days (18 days in the setter and 3 days in the hatchery). Finally, chicks aged 1 day were individually weighed and wing banded.
The chickens were placed within a brooder with incandescent lighting (23 h light:1 h darkness) on a litter floor. From 4 weeks of age, the chickens were grouped according to sex and then moved to a slat-floored barn with fluorescent lighting (23 h light:1 h darkness) until age 16 weeks. Fresh water and feed were provided ad libitum. The feed formulation, which excluded antimicrobial agents, was based on the method of Putsakul et al. (2010).
At age 16 weeks, approximately three birds from a full-sib family were randomly sampled to determine carcass characteristics and meat quality traits. The slaughtering process for ante-mortem and post-mortem followed good manufacturing practices for a poultry abattoir (Thai Agricultural Standard, 2005); carcass traits were dissected by cutting parts according to the standards for chicken meat (Thai Agricultural Standard, 2006). The performance data for the two progeny flocks were measured using the following parameters at age 16 weeks: BW, live body weight (LW), CW, BMW, LEG), wing weight (WW), cooking loss percentage (CL), drip loss percentage (DL), shear force value (SF), lightness (L*), redness (a*), and yellowness (b*).
The left side of the breast muscle was weighed, placed in a plastic bag, hung on the shelf, and stored at 4°C for 24 h. Subsequently, each sample was then removed from the plastic bag, excess surface moisture was wiped off, and the sample was re-weighed to determine the DL values (Northcutt et al., 1994). The breast meat colors, L*, a*, and b*, were measured using a Hunter Lab colorimeter (Reston, Virginia, USA) at 24 h post-mortem according to the CIE (Commission International de I'Eclairage) method (International Commission on Illumination, 1978).
Cooking loss of the right breast muscle was measured following the method of Papinaho and Fletcher (1996). The sample was weighed and cooked for 15 min at 95°C in steam, cooled to room temperature for 20 min, and then re-weighed. For the SF, the sample was cut to 2.0 × 1.0 × 1.5 cm (Saláková et al., 2009) and analyzed using a Lloyd Texture analyzer with a Warner-Bratzler blade attachment (Bognor Regis, West Sussex, UK).
2.3 Genotypic data
A blood sample was collected from the jugular vein during the slaughtering process. Genomic DNA was isolated using the alkaline lysis method (Sambrook et al., 1989). The PCR primer pairs are shown in Table 1 according to Okumura et al. (2006). The PCR conditions of CAPN1 were initial denaturation at 94°C for 5 min, 35 cycles of denaturing at 94°C for 30 s, annealing at 65°C for 30 s, extension at 72°C for 30 s, and final extension at 72°C for 10 min. The PCR samples of CAPN1 were run on 4.5% polyacrylamide gel at the DNA Technology Laboratory, Kamphaeng Saen, Thailand. Subsequently, the homozygous individuals were sequenced using a 3500xL Genetic Analyzer (Applied Biosystems, USA). Nucleotide sequences were annotated using a nucleotide BLAST program from the National Center for Biotechnology Information (NCBI). Differences in the homologous genotypes were aligned using the Clustal X software (Larkin et al., 2007) and were analyzed using GeneDoc program (Nicholas & Nicholas, 1997).
The PCR conditions for CAPN3 were initial denaturation at 95°C for 5 min, 30 cycles of denaturing at 95°C for 30 s, annealing at 68°C for 30 s, extension at 72°C for 3 min, and final extension at 72°C for 5 min. Restriction fragment length polymorphism (RFLP) was used to identify the CAPN3 polymorphisms. The PCR product of the CAPN3 (5 μL) was digested with 10 units of HhaI enzyme (New England Biolabs Inc., USA) at 37°C for 4 h. After digestion, the DNA fragments were separated on a 1% agarose gel using electrophoresis. The gel was stained using the SERVA DNA Stain G (SERVA, Germany) and visualized under ultraviolet light.
2.4 Statistical analysis
All statistical testing, including the genotypic and allelic frequencies for each locus based on the PROC FREQ procedure, was carried out using the SAS® on Demand for Academics (SAS Institute Inc., Cary, North Carolina, USA). Hardy–Weinberg equilibrium was tested using the chi-square test () based on the method of Falconer and Mackay (1996) The genotypic and allelic substitution effects of the CAPN1 and CAPN3 were analyzed using the PROC GLM procedure, while least squares means (LSM) and the comparison of the LSM of the genotypic effect of those genes were analyzed using Tukey's test in the SAS® on Demand for Academics (SAS Institute Inc., Cary, North Carolina, USA). The model was yijklm = μ + Hi + Sj + G1k + G2l + G1k * G2l + eijklm, where yijklm is the individual performance BW, LW, CW, BMW, LEG, WW, CL, DL, SF, L*, a*, and b*; μ is the overall mean; Hi is the fixed effect of hatching batch i (i = 1 and 2); Sj is the fixed effect of sex j (j = male and female); G1k is the fixed effect of the CAPN1 genotypes k (k = AA, AB, and BB); G2l is the fixed effect of the CAPN3 genotypes l (l = CC, CT, and TT); G1k * G2l is the interaction between the CAPN1 and CAPN3 genotypes; and eijklm is the random residual effect ~ NID (0, ).
The regression coefficients were estimated to determine the allele substitution effects of the CAPN1 and CAPN3. The fixed genotypic effects were replaced by coding 0, 1, and 2 for the BB, AB, and AA genotypes of the CAPN1 or the TT, CT, and CC genotypes of the CAPN3, respectively. The significance of the allele substitution effect was examined using the PROC GLM procedure.
3 RESULTS
3.1 Performance of growth, carcass, and meat quality traits
The descriptive statistics are shown in Table 2 for growth (BW and LW), carcass yield (CW, BMW, LEG, and WW), meat quality (CL, DL, and SF), and meat color (L*, a*, and b*) in Betong chicken (KU line). The chickens reached a BW of 1939.15 g, which is close to the preferred market size (2 kg) at 16 weeks of age. Furthermore, the mean BMW in the current study was 275.55 g. The mean DL in the current study was 2.32%. Meat color was described by the values of L*, a*, and b* (62.01, 5.57, and 29.80, respectively). The mean of SF was 29.80 N (Table 2).
| Trait† | N | Mean ± SD‡ | Minimum | Maximum |
|---|---|---|---|---|
| BW | 534 | 1939.15 ± 374.03 | 1224.00 | 2,949.00 |
| LW | 251 | 1912.49 ± 374.65 | 1197.00 | 2,763.00 |
| CW | 252 | 1557.48 ± 330.78 | 960.00 | 2,235.00 |
| BMW | 252 | 275.55 ± 42.98 | 159.00 | 399.00 |
| LEG | 252 | 492.03 ± 121.24 | 266.00 | 738.00 |
| WW | 252 | 172.88 ± 34.88 | 113.00 | 262.00 |
| CL | 252 | 23.95 ± 3.62 | 12.36 | 42.72 |
| DL | 249 | 2.32 ± 0.71 | 0.77 | 4.35 |
| SF | 252 | 29.80 ± 12.31 | 14.98 | 90.92 |
| L* | 251 | 62.01 ± 2.82 | 51.57 | 67.63 |
| a* | 251 | 5.57 ± 0.95 | 3.30 | 9.47 |
| b* | 251 | 17.60 ± 2.47 | 11.18 | 23.06 |
- † BW = body weight at age 16 weeks (g); LW = live weight (g); CW = carcass weight (g); BMW = breast meat weight (g); LEG = leg weight (g); WW = wing weight (g); CL = cooking loss (%); DL = drip loss (%); SF = shear force (N); L* = lightness; a* = redness and b* = yellowness.
- ‡SD = standard deviation.
3.2 Genotypic polymorphisms of CAPN1 and CAPN3
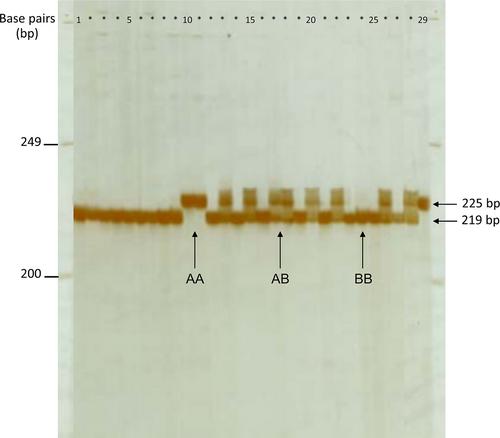
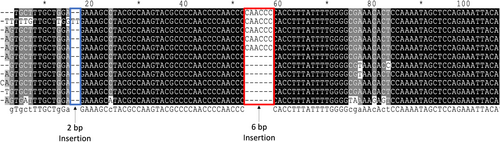
The blood samples were extracted for DNA analysis. Fragments of the CAPN1 were amplified and denatured using PCR and were visible in a 4.5% polyacrylamide gel. The CAPN1 polymorphisms were classified into three genotype groups: AA, AB, and BB (Figure 1). The product sizes of the AA and BB genotypes were 225 and 219 bp, respectively, whereas the product sizes of the AB genotype covered both 225 and 219 bp. The DNA sequences of all genotypes are shown in Figure 2. The difference between the AA and BB genotypes was caused by a 6-bp single insertion (CAACCC). From DNA sequencing, a 2-bp (TT) insertion was found in a sample, as shown in line 2 in Figure 2.
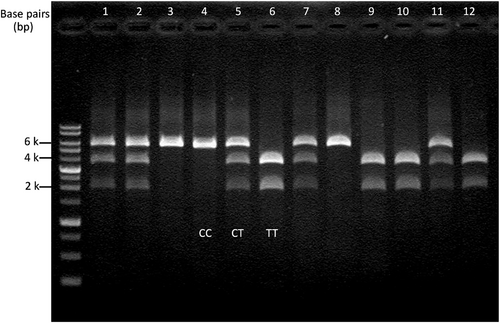
Polymorphisms of the CAPN3 were found in this population based on PCR-RFLP. Consequently, three genotypes were identified: CC, TT, and CT. The CC genotype presented a single 6-kbp fragment, whereas the TT genotype presented two fragments of 2 and 4 kbp. The heterozygous genotype CT had three fragments of 2, 4, and 6 kbp (Figure 3).
The allelic and genotypic frequencies of the CAPN1 and CAPN3 are shown in Table 3. For the CAPN1, the allelic frequency of the A allele (0.22) was lower than that of the B allele (0.78). Consequently, the genotypic frequency of AA (0.05) was lower than that of BB (0.61) and the frequency of AB was 0.34. On the other hand, for the CAPN3, the allelic frequencies of C (0.54) and T (0.46) were found in nearly half the occurrences. Therefore, the striking genotypic frequency was the CT genotype (0.50), while the genotypic frequencies of CC and TT were 0.29 and 0.21. Moreover, the genotypic frequencies of CAPN1 and CAPN3 were in Hardy–Weinberg equilibrium.
| Gene | Genotypic frequency | Allelic frequency | ||||
|---|---|---|---|---|---|---|
| CAPN1 | AA | AB | BB | A | B | |
| 0.05 | 0.34 | 0.61 | 0.22 | 0.78 | 0.0063 | |
| CAPN3 | CC | CT | TT | C | T | |
| 0.29 | 0.50 | 0.21 | 0.54 | 0.46 | 0.0066 | |
- n = 252, = 5.99.
3.3 Genotypic effects of the CAPN1 and CAPN3 on growth, carcass, and meat quality traits
The genotypic effects of CAPN1 and CAPN3 with the interaction between the genes for all traits were analyzed, and the interactions were not significant except for the L* trait (Table 4). The LSM of the growth, carcass, and meat quality traits for each of the CAPN1 and CAPN3 genotypes are shown in Table 5. The genotypic effects of CAPN1 were not significant for all traits, whereas the genotypic effect of the CAPN3 showed association with BMW. The TT genotype was more favorable than the other genotypes for BMW (P < 0.05).
| Trait† | P-value |
|---|---|
| BW | 0.5707 |
| LW | 0.8386 |
| CW | 0.8460 |
| BMW | 0.7700 |
| LEG | 0.8644 |
| WW | 0.9812 |
| CL | 0.9376 |
| DL | 0.7626 |
| SF | 0.7330 |
| L* | 0.0106 |
| a* | 0.4363 |
| b* | 0.4772 |
- †BW = body weight at age 16 weeks (g); LW = live weight (g); CW = carcass weight (g); BMW = breast meat weight (g); WW = wing weight (g); LEG = leg weight (g); CL = cooking loss (%); DL = drip loss (%); SF = shear force (N); L* = lightness; a* = redness and b* = yellowness.
| Trait† | Genotype of CAPN1‡ | Genotype of CAPN3‡ | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | P-value | CC | CT | TT | P-value | |||||||
| LSM | SE | LSM | SE | LSM | SE | LSM | SE | LSM | SE | LSM | SE | |||
| BW | 1,993.00 | 51.51 | 1,979.73 | 18.77 | 1,963.45 | 14.38 | 0.7146 | 1,941.94 | 28.35 | 1,955.80 | 26.44 | 2,038.43 | 41.41 | 0.1428 |
| LW | 1,932.57 | 47.22 | 1,916.04 | 17.07 | 1,906.03 | 13.14 | 0.8032 | 1,875.58 | 25.96 | 1,898.26 | 24.24 | 1,980.79 | 37.90 | 0.0708 |
| CW | 1,579.20 | 39.02 | 1,556.68 | 14.10 | 1,550.30 | 10.76 | 0.7520 | 1,534.99 | 21.45 | 1,547.14 | 20.03 | 1,604.05 | 31.30 | 0.1804 |
| BMW | 287.23 | 9.09 | 277.08 | 3.29 | 277.13 | 2.51 | 0.5540 | 273.11b | 5.00 | 273.41b | 4.67 | 294.92a | 7.29 | 0.0282 |
| LEG | 505.50 | 14.06 | 491.03 | 5.08 | 489.06 | 3.88 | 0.5266 | 487.08 | 7.73 | 492.68 | 7.22 | 505.83 | 11.28 | 0.3921 |
| WW | 172.77 | 4.36 | 170.55 | 1.58 | 173.08 | 1.20 | 0.4413 | 169.22 | 2.40 | 171.74 | 2.24 | 175.44 | 3.50 | 0.3375 |
| CL | 24.06 | 1.09 | 23.06 | 0.39 | 24.18 | 0.30 | 0.5552 | 23.79 | 0.60 | 24.27 | 0.56 | 23.82 | 0.88 | 0.8240 |
| DL | 2.30 | 0.18 | 2.35 | 0.06 | 2.28 | 0.05 | 0.6950 | 2.21 | 0.10 | 2.48 | 0.09 | 2.23 | 0.14 | 0.0898 |
| SF | 27.79 | 3.81 | 28.15 | 1.38 | 30.19 | 1.05 | 0.4572 | 28.52 | 2.09 | 30.04 | 1.96 | 27.58 | 3.05 | 0.7597 |
| L* | 60.14 | 0.87 | 61.83 | 0.31 | 62.26 | 0.24 | 0.0522 | 61.73 | 0.48 | 61.86 | 0.45 | 60.65 | 0.70 | 0.3258 |
| a* | 5.66 | 0.30 | 5.63 | 0.11 | 5.53 | 0.08 | 0.7144 | 5.69 | 0.17 | 5.68 | 0.15 | 5.46 | 0.24 | 0.7020 |
| b* | 17.00 | 0.67 | 17.70 | 0.24 | 17.65 | 0.18 | 0.6121 | 17.20 | 0.37 | 17.82 | 0.34 | 17.33 | 0.54 | 0.4369 |
- †BW = body weight at age 16 weeks (g); LW = live body weight (g); CW = carcass weight (g); BMW = breast meat weight (g); LEG = leg weight (g); WW = wing weight (g); CL = cooking loss (%); DL = drip loss (%); SF = shear force (N); L* = lightness; a* = redness and b* = yellowness.
- ‡Least squares means within a row with different superscript letters differ from adjusted Tukey's test (P < 0.05).
Regardless of the interaction between the genotypic effect of the genes, the allele substitution effect of the CAPN1 had a significant association with L* (P < 0.05) (Table 6). Substitution of the B allele with the A allele led a reduction in the L* value in meat (−0.78 ± 0.30). Furthermore, the allele substitution effect of the CAPN3 was significantly associated with BW, LW, CW, and BMW (P < 0.05). Substitution of the T allele with the C allele reduced BW, LW, CW, and BMW, accounting for −40.58 ± 14.77, −44.53 ± 13.39, −25.14 ± 11.02, and −8.63 ± 2.61 g, respectively (Table 6).
| Trait† | N | CAPN1 | P-value | CAPN3 | P-value | ||
|---|---|---|---|---|---|---|---|
| Allele substitution | Allele substitution | ||||||
| Effect | SE | Effect | SE | ||||
| BW | 249 | 6.84 | 17.56 | 0.6973 | −40.58 | 14.77 | 0.0065 |
| LW | 251 | 5.77 | 16.00 | 0.7189 | −44.53 | 13.39 | 0.0010 |
| CW | 252 | 5.30 | 13.21 | 0.6887 | −25.14 | 11.02 | 0.0234 |
| BMW | 252 | 2.51 | 3.12 | 0.4522 | −8.63 | 2.61 | 0.0011 |
| LEG | 252 | 3.77 | 4.76 | 0.4286 | −7.56 | 3.97 | 0.0580 |
| WW | 252 | −0.85 | 0.76 | 0.2634 | −0.49 | 0.63 | 0.4382 |
| CL | 252 | −0.25 | 0.37 | 0.5046 | −0.12 | 0.31 | 0.7044 |
| DL | 249 | 0.04 | 0.06 | 0.5173 | −0.04 | 0.05 | 0.4383 |
| SF | 252 | −1.28 | 1.30 | 0.3255 | −0.62 | 1.08 | 0.5699 |
| L* | 251 | −0.78 | 0.30 | 0.0109 | −0.03 | 0.25 | 0.9054 |
| a* | 251 | 0.16 | 0.10 | 0.1109 | 0.04 | 0.09 | 0.6817 |
| b* | 251 | −0.05 | 0.23 | 0.8426 | −0.36 | 0.19 | 0.0565 |
- †BW = body weight at age 16 weeks (g); LW = live weight (g); CW = carcass weight (g); BMW = breast meat weight (g); LEG = leg weight (g); WW = wing weight (g); CL = cooking loss (%); DL = drip loss (%); SF = shear force (N); L* = lightness; a* = redness and b* = yellowness.
4 DISCUSSIONS
The chickens could reach a BW of 1.94 kg that was close to the preferred market size (2 kg) at age 16 weeks (Table 2) and was consistent with other research using the Betong chickens (KU line) (Putsakul et al., 2010; Wangtaweesukkamol et al., 2013). Furthermore, the mean carcass traits (CW, BMW, LEG, and WW) in the current study were higher than previous reports for Betong chicken (Gongruttananun & Chotesangasa, 1996) and black chicken lines (Sungkhapreecha et al., 2015). Likewise, the BMW and LEG percentages in the current study were higher than another study (Makchumpon et al., 2015) (Table 2). The mean DL in the current study was 2.32%, which was consistent with the normal standard (less than 3%) (Honikel & Hamm, 1994). The values of L*, a*, and b* were higher than those previously reported for other native chickens (Chabault et al., 2012; Wattanachant et al., 2004). The average of SF was different from other reports in native chickens (Jaturasitha et al., 2002, 2008) (Table 2). The SF values were varied depending on several factors such as sex, age, and strain of chickens.
In chicken, the CAPN1 is located on chromosome 3 (Okumura et al., 2005; Sorimachi et al., 1994). In the current study, for the polymorphisms of CAPN1, the lengths of two bands (225 and 219 bp), which was confirmed by sequencing on BLAST search in the NCBI data, differed from 261 and 255 bp as reported by Okumura et al. (2006) in an East Lansing reference population and Kobe University population. Moreover, the 6-bp single insertion was CAACCC, as shown in Figure 1, whereas Okumura et al. (2005) found CCCCAA insertion. The nucleotide sequence of the CAPN1 in intron 4 was confirmed with the GenBank accession number of AB177603.1. However, Okumura et al. (2005) could not find CAPN1 in a specific genomic position by using the BLAST search.
The CAPN3 is located on chromosome 5, with a total length of 6 kbp in exons 4–11 (Okumura et al., 2005). Due to the large size of the amplified product, CAPN3 could not be sequenced in this study. Therefore, the reference FASTA file for CAPN3 from NCBI database (Genebank accession number 423233) was used. The reference sequence was then cut with the restriction enzyme HhaI using SnapGene® software. The recognition sequence for HhaI is GCG/C (New England Biolabs, USA). Alignment with genome database (Ensembl: ENSGALG00010019472) revealed that the SNP is located within intron 6 of CAPN3.
The polymorphisms of CAPN3 had similar patterns to the results reported by Okumura et al. (2006). The allelic and genotypic frequencies of CAPN1 and CAPN3 in this population were in Hardy–Weinberg equilibrium, indicating that the cocks and hens were randomly sampled.
Even though the genotypic effects of CAPN1 on growth, carcass, and meat quality in poultry were not significant in the current study (Table 5), the significant allele substitution effect of CAPN1 was found for L* (Table 6) and the combination of CAPN1 and CAPN3 could improve L* (Table 4). Previous studies reported the associations among the SNPs of CAPN1 and carcass and meat quality in poultry (Rasouli et al., 2013; Zhang et al., 2008). Correlations between CAPN1 and b*, as well as between CAPN1 and SF, have been reported in Japanese quail (Rasouli et al., 2013). Furthermore, the haplotypes of CAPN1 that were produced from mutation (SNPs; 2546C > T, 3535G > A, and 7198C > A) were associated with carcass and muscle fiber in Chinese local chicken breeds. However, Zhang et al. (2008) showed that the CAPN1 had no significant effects for all performance traits. Therefore, the effect of CAPN1 has been found effective in some meat quality traits in some populations.
Besides, the effects of CAPN3 genotypes on growth, carcass, and meat quality were not significant except for BMW and the allele substitution effect of the CAPN3 was significantly associated with BW, LW, CW, and BMW (Tables 5 and 6). Likewise, Zhang et al. (2009) reported that the SNP haplotypes of CAPN3 (11,818 T > A and 12,814 T > G) were associated with some carcass traits (BW, CW, BMW, and LEG) in five commercial pure lines and four native breeds in China. Furthermore, the polymorphisms of CAPN3 have been found to be related to thigh yield, thawing-cooking loss, and SF in broilers in Brazil (Felício et al., 2013).
Even though CAPN1 and CAPN3 are indirectly involved in muscle cell metabolism, their genotypes are related to meat quality. Calpain1 is active in the Z-line, while calpain3 is found in the sarcomere near the Z and M-lines of muscle fibers (Ilian et al., 2004; Koohmaraie, 1994). Thai native chickens, including Betong chickens (KU line), are slow-growing breeds. Piórkowska et al. (2015) reported that fast-growing broiler chickens had higher expression levels of CAPN1 and CAPN3 in breast muscle compared to slow-growing broiler chickens. Associations between CAPN1 polymorphisms and live weights were found in slow-growing chicken breeds (Kubota et al., 2019). Negative correlation between the CAPN1 expression level and the shear force of breast muscle in fast-growing female chickens was found. Calpain3 was necessary for normal muscle tissue homeostasis and function (Piórkowska et al., 2015) and was strongly dependent on calcium release and content in skeletal muscle fibers (Kramerova et al., 2008).
The two forms of calpain, particularly μ-calpain and μ/m-calpain, are specific to birds and are both calcium-sensitive. The μ/m-calpain remained very stable by 24 h postmortem, whereas μ-calpain (calpain1) levels had strongly decreased. This difference in stability explained why avian muscle tenderizes rapidly (Lee et al., 2008). An association between the genotype of CAPN1 and meat lightness was found in this study. This could be explained that caplain1 broke down muscle protein like actin and myosin after slaughter and the unfolded proteins made the meat appear lighter (Kemp et al., 2010). Moreover, the genotype of CAPN3 were found to be associated with body and CW. This confirmed the participation of calpain3 in regulation of myogenesis. Furthermore, Zhang et al. (2012) showed that the expression of the CAPN3 mRNA was related to muscle fiber development.
In conclusion, the polymorphisms of CAPN1 and CAPN3 were found in the Betong chicken (KU line) population. The CAPN3 was suitable as a marker gene for BW, LW, CW, and BMW. Betong (KU line) chickens with the TT genotype of CAPN3 obtained the greatest advantage for growth and some carcass yields, especially for LW and BMW. The CAPN1 genotype as well as the combination of the CAPN1 and CAPN3 genotypes could be used for genetic markers for L*. Incorporating molecular markers for these genes into the selection process could improve genetic gain for growth, carcass yield, and the lightness of meat color in this population.
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid for Scientific Research from Kasetsart University Scholarship for Doctoral Students (Grant no. 5410103). Special thanks to Assoc. Prof. Dr. Chatchawan Jantasuriyarat, Department of Genetics, Faculty of Science, Kasetsart University, Bangkok, Thailand, for advice and guidance regarding molecular information and to Dr. Melody Muguerza for reviewing and editing an earlier draft of the manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.




