HCM-associated ALMS1 variant: Allele drop-out and frequency in Italian Sphynx cats
Abstract
Hypertrophic cardiomyopathy (HCM) is the most common cardiomyopathy in domestic cats, and some inherited variants are available for genetic testing. A variant of the Alstrom syndrome protein 1 gene (ALMS1) was recently reported to be associated with HCM in the Sphynx cat breed (A3: g.92439157G>C). Genetic screening of the variant, promoted by the Osservatorio Veterinario Italiano Cardiopatie and Genefast Laboratory, was offered to Sphynx cat owners and breeders in Italy. Genotype data were initially obtained by Sanger sequencing. In one case where the samples of a trio were available, inconsistency in the vertical transmission of the variant suggested an allele dropout (ADO) of the wt allele. A new external primer pair was designed as an alternative to the original. The larger PCR product obtained was sanger sequenced, and five novel single nucleotide variants (SNVs) not yet annotated in open-access databases were detected. Three of these SNVs were within the original primer-binding regions and were assumed to have caused ADO. The haplotype, including the ADO SNVs, was detected in two cats belonging to different lineages. To accurately genotype ALMS1 g.92439157G>C in the samples, we set up a real-time TaqMan MGB assay while avoiding all surrounding SNVs. At g.92439157G>C, for 136 Sphynx cats, g.92439157 C variant was highly widespread (freq. >0.50). The present study reports five new variants surrounding ALMS1 g.92439157G>C that must be considered when designing the test. The study also indicates the need to verify the correspondence between the g.92439157 C variant frequency and the prevalence of HCM by increasing clinical visits and follow-ups and finally to promote genetic counselling for accurate management of mating plans in Italian Sphynx cats.
Hypertrophic cardiomyopathy (HCM) is the most common cardiomyopathy in domestic cats. In pure breeds and randomly bred cats, causative variants of HCM have been identified in different genes (Meurs et al., 2005, 2007; Schipper et al., 2019) and diagnostic genetic tests are widely used to help practitioners with prognosis and breeders with selection. Recently, a single nucleotide variant (SNV) in the Alstrom syndrome protein 1 gene (ALMS1) was reported to be associated with HCM in the Sphynx cat breed, resulting in an immediate request for testing by veterinarians, owners and breeders. The variant is a G>C substitution in ALMS1 exon 12 at the feline genome position A3: 92439157 (ENSFCAG00000008756, vs.9), which is predicted to change glycine to arginine (Meurs et al., 2021).
In 2021, 43 male and 69 female Sphynx cats were registered with the Associazione Nazionale Felina Italiana, the biggest fancy cat association in Italy. Soon after the identification of the ALMS1 variant, the authors, via the Osservatorio Veterinario Italiano Cardiopatie (OVIC) and partner laboratory Genefast, promoted genetic screening of the variant at the national level for owners and breeders. Samples were also collected from customers who were not affiliated with the OVIC and did not request echocardiography. Primers ALMS1_F and ALMS1_R were designed based on the Felis catus genomic sequence vs.9 (Buckley et al., 2020) to obtain a 157 bp amplicon and genotyping was performed using Sanger sequencing (Figure 1).
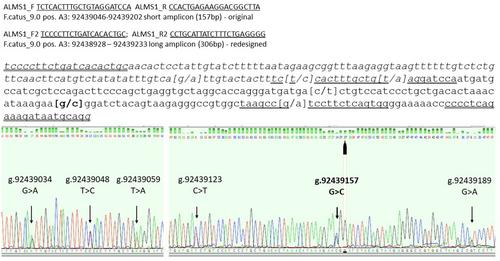
An inconsistency between a homozygous variant sire and his homozygous wt offspring was reported by a breeder. The breeder provided a blood sample from the dam that tested heterozygous. After confirming the parental relationships using the ISAG STR panel, allele dropout (ADO) of the wt allele in the father was hypothesised. A new set of PCR primers were then designed, covering the surrounding region of the ALMS1 variant and the original PCR primer sites. The new primers (ALMS1_F2 and ALMS1_R2) generated a 306 bp amplicon (Figure 1; PCR protocol in Table S1). Using 19 selected samples, including the trio and 12 g.92439157G>C variant homozygotes, redesigned PCRs and Sanger sequencing were performed. Within the trio, the sire was heterozygous, confirming that a wt allele dropout had previously occurred when the original PCR assay was used. By aligning all longer sequences, five SNVs other than the published g.92439157G>C that were not already annotated in public databases were detected: g.92439034G>A; g.92439048T>C; g.92439059T>A; g.92439123C>T; and g.92439189G>A (Figure 1). Two (g.92439048T>C and g.92439059T>A) were in the forward (ALMS1_F) and one (g.92439189G>A) was in the reverse (ALMS1_R) primer site of the original short PCR; these are very probably the cause of ADO in the sire (Figure 1). Among the cats originally genotyped as homozygous for the g.92439157 C variant, there was another cat in addition to the sire of the trio that yielded a heterozygous result with the new primer set. Importantly, the two cats with ADO were not related based on their pedigree.
The five novel SNVs were either intronic substitution – g.92439034G>A, g.92439048T>C and g.92439059T>A – or exonic synonymous substitution – g.92439123C>T and g.92439189G>A (Figure 1). Two intronic SNV (g.92439048T>C and g.92439059T>A) were clustered near the splicing site but were not likely to cause alternative splicing.
Haplotypes were inferred using chromatograms of either the wt or var homozygote samples at the g.92439157G>C position and trio samples. Three different haplotypes were identified: GTTTCG associated with the ALMS1 g.92439157 C variant (underlined) and ACTCGG and ACACGA, both associated with the g.92439157 G wt nucleotide (underlined); the ACACGA haplotype was recorded in the ADO cases (Table S2). Excluding the offspring in the trio, in 18 unrelated samples 100% of the g.92439157 C alleles showed the GTTTCG haplotype, whereas the g.92439157 G variant was associated 75% with the ACTCGG haplotype and 25% with the ACACGA haplotype.
To accurately genotype the g.92439157G>C variation in all samples, avoiding all SNVs in the flanking regions, a real-time TaqMan MGB assay with primers F-sphynx CCATCCCTGCTGACACTAAACAT and R-sphynx GGTTTTTCCCCACTGAGAAGGA and G allele probe (wt) VIC-CTTACTGTAGATCCCTTCTTT-NFQ and C allele probe (variant) FAM-CTGTAGATCCGTTCTTT-NFQ was designed. The PCR mix contained 1× TaqManUniversal PCR Master Mix (Thermo Fisher), 1× Custom Taqman SNP Genotyping Assay (Thermo Fisher), 2.5 μl of DNA template, and molecular biology-grade water to a final volume of 25 μl. Each sample was assayed in duplicate and no DNA control was assayed in each run. The assay was validated using all the collected samples.
Of the 136 Sphynx cat sampled, with or without clinical data, the genotype frequencies were distributed as follows: 33 wt/wt (24.3%), 67 wt/var (49.3%) and 36 var/var (26.5%), and the var allele frequency was 51.1%. Considering only the younger cats (n. 59, ≤4 years old), the genotypes were distributed as follows: 18 wt/wt (30.5%), 27 wt/var (45.8%) and 14 var/var (23.7%), and a var allele frequency of 46.6%, showing widespread presence of the variant in the younger generations of pet and breeding cats.
In the last decade, the number of genetic variants identified as causative of diseases has greatly increased in pets, and diagnostic tests are now widely used, helping practitioners in diagnosis and prognosis and breeders in selection. Good interactions between owners, breeders, breeders’ associations, practitioners, researchers and service laboratories (as in OVIC) can be functional and beneficial for monitoring genetic frequencies and disease prevalence in a region. Allele dropout artefacts are caused by occasional amplification failure of one of the two alleles at a given genomic DNA locus. It represents a well-known risk of misdiagnosis, particularly for amplification-based methods (0.44% occurrence of genotype mistypings in humans) (Blais et al., 2015) and reduces the diagnostic yield of genetic testing in cardiomyopathies (Shestak et al., 2021). However, in humans within PCR failure cases systematic allele-specific failures owing to sequence variants occur in only 6% of cases (95% CI, 3%–11%) (Blais et al., 2015). Of these, 75% variants were in the primer-binding sites, whereas the remaining were located within the amplicon (Blais et al., 2015). However, when comparing the percentages, it should be noted that the studies and annotations available are still more extensive for humans than for cats. In the present study, by re-genotyping 12 g.92439157 C homozygotes, we detected SNVs in the binding sites of the original primer pair set, identifying a non-random ADO of the wt allele in two unrelated cats. If the ADO was mainly due to the absence of annotation of those variants into public variant repositories, which misled the first genotyping design, the high proportion of ADO in such a small sample could be motivated by high inbreeding and thus genetic similarity in the Sphynx population. The authors did not find evidence to explain why the two ADO cases, which also shared the ACACGA haplotype, had no kinship ties based on their pedigree. This could have been due to misregistration or the presence of a common ancestor several generations prior. Despite having solved with a new PCR design the risk of ADO of the wt allele and to overestimate the variant allele, the authors recorded a percentage of the g.92439157 C variant above 45%, which is high for a causative variant, even considering possible inflation by some unavoidable collection bias. Echocardiographic controls were implemented by the Osservatorio, and 75.3% of the healthy Sphynx cats that underwent echocardiography showed the g.92439157 C variant in a single or double dose. However, these controls do not permit the evaluation of the effect of the g.92439157 C variant on the disease in the Italian Sphynx population, owing to low numerosity and the fact that only a relatively small number of cats had reached an age of approximately 4 years, at which one can be definitively confident in the phenotype classification.
Overall, the present data indicate that care must be taken with good practice (CLSI, 2012) when typing the ALMS1 g.92439157 C variant. The authors suggest the need for close genetic counselling of the breeders, to avoid dramatic reduction of the genetic pool owing to uncontrolled genetic testing and mating exclusion, and the need to improve clinical controls to monitor the effect of the variant on the health of the Italian population.
ACKNOWLEDGEMENTS
The authors thank Animal Bio Arkivi at the University of Milan, all of the specialists of the Osservatorio Veterinario Italiano Cardiopatie, the Associazione Nazionale Felina Italiana Studbook, Mr Vincenzo Macrì, and the Italian Sphynx breeders and owners for fruitful collaboration.
CONFLICT OF INTEREST STATEMENT
The authors have no conflicts of interest to declare.
Open Research
DATA AVAILABILITY STATEMENT
The five SNVs identified in this study have been deposited in the European Variation Archive (Cezard et al., 2021) at EMBL-EBI under accession no. PRJEB60498 (https://www.ebi.ac.uk/eva/?eva-study=PRJEB60498).




