The prion-related protein (testis-specific) gene (PRNT) is highly polymorphic in Portuguese sheep
Summary
The objective of this study was to search for polymorphisms in the ovine prion-related protein (testis-specific) gene (PRNT). Sampling included 567 sheep from eight Portuguese breeds. The PRNT gene-coding region was analyzed by single-strand conformation polymorphism and sequencing, allowing the identification of the first ovine PRNT polymorphisms, in codons 6, 38, 43 and 48: c.17C>T (p.Ser6Phe, which disrupts a consensus arginine-X-X-serine/threonine motif); c.112G>C (p.Gly38>Arg); c.129T>C and c.144A>G (synonymous) respectively. Polymorphisms in codons 6, 38 and 48 occur simultaneously in 50.6% of the animals, 38.8% presenting as heterozygous. To study the distribution of the polymorphism in codon 43, a restriction fragment length polymorphism analysis was performed. Polymorphic variant c.129C, identified in 89.8% of the animals with 32.8% presented as heterozygous, was considered the wild genotype in Portuguese sheep. Eight different haplotypes which have comparable distribution in all breeds were identified for the PRNT gene. In conclusion, the PRNT coding region is highly polymorphic in sheep, unlike the prion protein 2 dublet gene (PRND), in which we previously found only one synonymous substitution (c.78G>A), in codon 26. The absence or reduced number of PRND heterozygotes (c.78G>A) was significantly associated with three PRNT haplotypes (17C-112G-129T-144A,17CT-112GC-129CT-144AG and 17T-112C-129C-144G), and the only three animals found homozygous at c.78A had the 17C-112G-129C-144A PRNT haplotype. These results constitute evidence of an association between polymorphic variation in PRND and PRNT genes, as has already been observed for PRND and prion protein gene (PRNP).
Transmissible spongiform encephalopathies, including scrapie disease in sheep, are fatal neurodegenerative disorders caused by a post-translational change in the conformation of cellular prion protein – PrPC – a membrane glycoprotein encoded by the prion protein (PRNP) gene into the pathogenic scrapie isoform PrPSc (Prusiner 1998). The PRNP gene and its homologs – the prion protein 2 (dublet) (PRND), shadow of prion protein homolog (zebrafish) (SPRN) and prion-related protein (testis-specific) (PRNT) genes – constitute the ‘prion gene complex’ (reviewed in Pimenta et al. 2011). PRNP, PRND and PRNT coding genes are located in close proximity in Ovis aries chromosome 13. PRND and PRNT are in opposite orientations, which is typical of genomic regions with similar structural organization and expression profiles. PRNT is located downstream to PRND and PRNP in sheep genomic sequences, closer to PRND (6866 bp) than to PRNP (20427 bp).
PRNP is highly expressed in the central nervous system and testis (Makrinou et al. 2002). Unlike PRNP, PRND is poorly expressed in the brain and the Doppel (Dpl) protein is not needed for prion disease progression (Behrens et al. 2001), but the apparent co-regulation of PrPC and Dpl suggests a functional link between them (Moore et al. 2001). PRND is strongly expressed in the adult testis of sheep (Espenes et al. 2006), on both Sertoli and germinal cells. Prnd 0/0 mice males are sterile (Behrens et al. 2002) and fertilizing ability of ovine spermatozoa (spz) increases after Dpl supplementation (Pimenta et al. 2012a). Prion testis-specific (Prt) protein is expressed in the testis during ram spermatogenesis (Pimenta et al. 2012b), and incubation of an anti-Prt antibody with ovine spz and oocytes reduced the number of fertilized oocytes and cleaved embryos (Pimenta et al. 2013). All these data strongly suggest a major physiological role for both Dpl and Prt in ram reproductive physiology.
The ovine PRNP gene-coding region is highly polymorphic. By contrast, only two synonymous substitutions (in codons 12 and 26) have been reported in the ovine PRND coding region (Comincini et al. 2001), besides the 26 SNPs deposited in dbSNP. Synonymous substitutions in codons 12 and 26 have not been associated with prion disease. In Portuguese sheep, we have only found the c.78C>G polymorphism, in codon 26, which showed an association with embryo production (Pereira et al. 2009) and semen traits/freezability (Baptista et al. 2008). Previously, we observed an association between the c.78C>G PRND polymorphism and PRNP genotypes that strongly affect the degree of susceptibility/resistance to scrapie in sheep (Mesquita et al. 2010), suggesting that the EU selection program to eradicate scrapie, based only on PRNP genotypes, may have unintended consequences on genetic diversity in sheep, with hypothetical repercussions on reproduction traits. Therefore, the main aim of the present work was to identify polymorphisms in the coding region of PRNT gene in sheep, also investigating putative associations between polymorphic variants of PRNT and the genotypes previously found in codon 26 of the PRND gene.
A total of 567 healthy animals (366 male, 201 female) from eight Portuguese sheep breeds were analyzed: 59 Bordaleira entre Douro e Minho (BordaleiraDM), 55 Churra Badana (CBadana), 131 Churra Galega Mirandesa (CGMirandesa), 16 Churra Mondegueira (CMondegueira), 57 Merino da Beira-Baixa (MBeiraBaixa), 59 Merino Branco (MBranco), 131 Saloia and 59 Serra da Estrela (SEstrela). We followed EU guidelines on farm animal welfare in accordance with EU Directive 86-609-EC and DL 129/92 of the Portuguese authority for animal experimentation.
Genomic DNA was extracted from sheep blood samples as in Mesquita et al. (2010). Specific primers to the PRNT coding region (forward: 5′-ATGGGTAGACAGAACTCTCCCA-3′; reverse: 5′-TTAGCAATAAATCTTTACTGACTCTCT-3′) were designed, based on the published Ovis aries PRNT mRNA sequence (NM_001097649.1). The PRNT and PRND coding regions were amplified and analyzed by single-strand conformation polymorphism (SSCP) as described in Pereira et al. (2009). After identifying the molecular nature and location of the polymorphisms (PRNT:c.144A>G, dbSNP accession 1850328053; PRNT:c.129T>C, accession 1850328054; PRNT:c.112G>C, accession 1850328055; PRNT:c.17C>T, accession 1850328056; PRND:c.78G>A, accession 1850328057), PRNT haplotypes were established. In order to study the distribution of one of the polymorphisms, found by sequencing but not easily related to the SSCP patterns obtained, a restriction fragment length polymorphism (RFLP) analysis was performed, using the restriction endonuclease SphI.
PRNT and PRND genotype frequencies were calculated, their distribution was compared in different populations, and deviations from Hardy–Weinberg equilibrium were evaluated using genepop software version 3.4 (Laboratoire de Genetique et Environment, Montpellier, France). Differences were considered significant when P < 0.05.
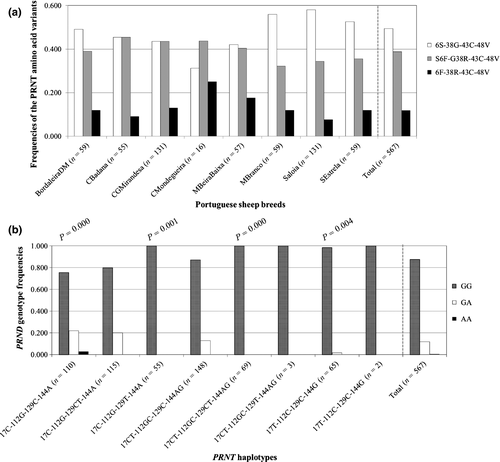
The 159-bp PCR fragment of the PRNT coding region was found to be highly polymorphic. Sequencing at first revealed three different haplotypes, resulting from three polymorphisms occurring simultaneously in codons 6, 38 and 48 of the PRNT gene: c.17C>T (p.Ser6Phe), c.112G>C (p.Gly38Arg) and c.144A>G (synonymous) respectively, when compared with the published ovine PRNT mRNA sequence (NM_001097649.1). Polymorphisms in codons 6, 38 and 48 occur simultaneously in 50.6% of the animals with 38.8% presented as heterozygous.
Sequencing revealed another synonymous polymorphism, in codon 43 (c.129T>C), which occurs independently from the other three and has been characterized through RFLP analysis. Polymorphic variant c.129C was identified in 89.8% of the animals with 32.8% presented as heterozygous and is considered the wild genotype, at least in Portuguese sheep.
Data on the frequencies of the eight PRNT haplotypes obtained for Portuguese sheep breeds are displayed in Table 1. Overall, the most frequent haplotype was 17CT-112GC-129C-144AG (26.1%), followed by 17C-112G-129CT-144A (20.3%) and 17C-112G-129C-144A (19.4%). The 17CT-112GC-129CT-144AG haplotype was more prevalent in CMondegueira (25%), CBadana (21.8%) and CGMirandesa (13.7%), and the 17T-112C-129C-144G haplotype in CMondegueira (25%) and MBeiraBaixa (17.5%). These last two haplotypes were the predominant ones in CMondegueira. The less frequent haplotypes were 17CT-112GC-129T-144AG (0.5%) and 17T-112C-129C-144G (0.4%); 17CT-112GC-129T-144AG was observed only in BordaleiraDM and SEstrela (3.4% and 1.7% respectively) and 17T-112C-129C-144G only in CGMirandesa (1.5%). There were no significant differences in the distribution of these haplotypes among the eight breeds. The eight breeds and the global population were not in Hardy–Weinberg equilibrium (χ2 = Infinity; df = 18; P = highly significant).
| Breed | n | PRNT haplotypes and aminoacid polymorphic variants (codons: 6–38–43–48) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 17C-112G-129C-144Aa | 17C-112G-129CT-144Aa | 17C-112G-129T-144Aa | 17CT-112GC-129C-144AGa | 17CT-112GC-129CT-144AGa | 17CT-112GC-129T-144AGa | 17T-112C-129C-144Ga | 17T-112C-129C-144Ga | ||
| 6S-38G-43C-48Vb | S6F-G38R-43C-48Vb | 6F-38R-43C-48Vb | |||||||
| BordaleiraDM | 59 | 0.186 | 0.237 | 0.068 | 0.186 | 0.169 | 0.034 | 0.119 | 0.000 |
| CBadana | 55 | 0.164 | 0.182 | 0.109 | 0.236 | 0.218 | 0.000 | 0.091 | 0.000 |
| CGMirandesa | 131 | 0.130 | 0.191 | 0.115 | 0.298 | 0.137 | 0.000 | 0.115 | 0.015 |
| CMondegueira | 16 | 0.063 | 0.188 | 0.063 | 0.188 | 0.250 | 0.000 | 0.250 | 0.000 |
| MBeiraBaixa | 57 | 0.193 | 0.123 | 0.105 | 0.298 | 0.105 | 0.000 | 0.175 | 0.000 |
| MBranco | 59 | 0.271 | 0.220 | 0.068 | 0.237 | 0.085 | 0.000 | 0.119 | 0.000 |
| Saloia | 131 | 0.221 | 0.244 | 0.115 | 0.260 | 0.084 | 0.000 | 0.076 | 0.000 |
| SEstrela | 59 | 0.271 | 0.186 | 0.068 | 0.288 | 0.051 | 0.017 | 0.119 | 0.000 |
| Total | 567 | 0.194 | 0.203 | 0.097 | 0.261 | 0.122 | 0.005 | 0.115 | 0.004 |
- n, number of animals analyzed in each breed.
- a PRNT haplotypes.
- b PRNT amino acid polymorphic variants.
The first four polymorphisms ever described in the ovine PRNT gene were revealed in this work, with two of them supporting changes in the Prt protein primary sequence. These could be of particular importance given that the role of Prt in ram spermatogenesis (Pimenta et al. 2012b) and in the initial steps of fertilization along the sperm-zona binding process (Pimenta et al. 2013) is now firmly established. The eight PRNT haplotypes give rise to three presumed major groups of amino acid polymorphic variants (Fig. 1a): 6S-38G-43C-48V (the most prevalent overall at 49.4%), S6F-G38R-43C-48V (at 38.8%) and 6F-38R-43C-48V (the least prevalent at 11.8%). Their distribution was different in the Saloia breed (P = 0.049) only when compared to CGMirandesa and CMondegueira. It is worth noting that the amino acid variant of serine at codon 6 (Ser6) always appears associated with the variant of glycine at codon 38 (Gly38), and in turn, phenylalanine (Phe6) always appears associated with arginine (Arg38), and finally, heterozygosity p.Ser6Phe always associates with heterozygosity p.Gly38Arg. Therefore, when studying animals characterized by one of the amino acid variants, the associated one should always be taken into account. The reported c.17C>T (p.Ser6Phe) substitution disrupts the consensus arginine-X-X-serine/threonine motif (where X represents any amino acid). O'Flaherty et al. (2004) previously reported that phosphorylation of the mentioned motif, characteristic of protein kinase A (PKA) substrates, is increased during human sperm capacitation. Furthermore, in human and mouse sperm, PKA has been found in both the acrosomal cap and the flagellum (Pariset & Weinman 1994; Visconti et al. 1997). Interestingly, Prt was detected by immunofluorescence at the acrosome region of ejaculated ram spz (Pimenta et al. 2012b). In the present work, we observed that the c.17C>T (Ser6) variant was the most common (Fig. 1a), followed by p.Ser6Phe and Phe6 variants. Taking all this into consideration, it is possible to infer that Prt might have a role in the phosphorylation signaling pathway, an important physiological prerequisite before the sperm cell can acrosome react and fertilize the oocyte.
Although some identified polymorphisms in the ovine PRND and PRNT genes are synonymous, recent studies have indicated that both synonymous and non-synonymous SNPs can influence mRNA stability, processing and maturation, thereby affecting its allelic expression (Komar 2007). Moreover, synonymous codon substitutions (namely changes from frequent to infrequent codons) may lead to different kinetics of mRNA (protein) translation and thus affect the co-translational folding pathway, yielding a protein with a different final structure and function (Komar 2007).
The distribution of PRND codon 26 polymorphic variants by PRNT haplotypes is represented in Fig. 1b. The population was in Hardy–Weinberg equilibrium (χ2 = 4.2; df = 8; P = 0.841). When compared with the total population, the absence of PRND heterozygotes (c.78G>A) was significantly associated with PRNT haplotypes 17C-112G-129T-144A (P = 0.001) and 17CT-112GC-129CT-144AG (P = 0.000). Additionally, of the 65 animals characterized with the 17T-112C-129C-144G haplotype, only one was heterozygous for PRND, which was significantly different from the global population (P = 0.004). Also, the only three animals found homozygous c.78A for PRND had the 17C-112G-129C-144A PRNT haplotype (P = 0.000). Thus, these animals are unique and should be conserved and further characterized.
In conclusion, data reported in this work reveal that PRNT is a highly polymorphic gene in Portuguese sheep. In addition, PRNT and PRND genotypes were associated, as previously observed between PRND and PRNP. These data strongly suggest that further studies are required to analyze the associations between these PRNT polymorphic variants and PRNP genotypes before implementing selection programs aiming to increase resistance to scrapie. As an important spermatogenesis-related protein, the fertility of the animals characterized by different PRNT genotypes and the alterations on protein structure for the p.Ser6Phe and p.Gly38Arg polymorphic variants should also be studied in detail in the near future.
Acknowledgements
This work has been supported by FCT – Fundação para a Ciência e a Tecnologia within the scope of the project PTDC/CVT/098607/2008.




