How alcohol makes the epigenetic clock tick faster and the clock reversing effect of abstinence
Tristan Zindler and Helge Frieling contributed equally.
Abstract
This study investigated the recently reported association between alcohol dependence and accelerated ageing and the potential effects of abstinence and relapse on DNA methylation status using Levine's epigenetic clock to estimate DNA methylation age in two independent cohorts. The first sample comprised 88 (15 female) detoxified patients with alcohol use disorder (AUD) and 32 (5 female) healthy control (CON) subjects (NCT02615977), and the second included 69 (10 female) AUD patients that were followed up for 12 months with respect to relapse (n = 38, 4 female) and abstinence (n = 31, 6 female) (NCT01679145). To account for the different aspects of ageing captured by various clocks, we performed additional analyses of the first-generation Horvath clock and next-generation Zhang clock. To account for the genetic liability of AUD and its potential influence on DNA methylation, we calculated a polygenic risk score for alcohol dependence. We found that ageing was accelerated by 3.64 years in AUD patients compared with the CON group according to Levine's DNAm PhenoAge. Furthermore, in a second longitudinal sample, we found that abstaining AUD patients displayed a decrease in DNAm PhenoAge by 3.1 years, but we found an over proportional increase by 2.7 years in those who relapsed. Polygenic risk did not affect epigenetic ageing within our sample. These results confirm the age acceleration associated with AUD and provide the first evidence for a recovery of this effect upon abstinence from alcohol.
1 INTRODUCTION
All multicellular organisms undergo an ageing process that is characterized by the gradual deterioration of biological functions, which is referred to as biological ageing in contrast to chronological ageing (corresponding to the time since birth).1 There are different options to assess biological ageing and its molecular and cellular correlates, for example, cell cycle arrest,2 telomere length in leukocytes,3 secretion of specific factors and cytokines,4 all providing important insights into the cellular signs of ageing. However, biomarkers used to measure cellular senescence are often unspecific and might fail to address the multi-level aspects of ageing. Epigenetic clocks provide a further valuable tool for monitoring the ageing process, because they result from changes in the organism at different levels. Deoxyribonucleic acid methylation (DNAm) clocks are comprehensive statistical models that use methylation levels at several specific cytosine and guanine dinucleotides (CpG) sites to calculate epigenetic age. There are two fundamental approaches to the methodology of DNAm clocks: The first aims to predict the chronological age of an individual as accurately as possible from DNAm (chronological DNAm clocks), whereas the second calculates biological age using other known indicators of biological ageing in addition to chronological age (biological DNAm clocks).5 The term ‘biological aging’ will subsequently be used with respect to DNA methylation estimates. Hence, aspects of ageing independent from methylation signals will not be covered by these estimates. While the molecular details of DNAm clocks are not well understood, they nonetheless enable investigations of the biological ageing process, which also reflects a person's health and wellbeing.6
Alcohol use disorder (AUD) is a chronic debilitating disorder associated with a reduced life expectancy due to an increased risk of mortality, limited treatment options and a poorly defined pathophysiology.7 Chronic exposure to alcohol over the lifespan has undisputed detrimental effects on health and well-being.8 It can therefore be assumed that AUD has an impact on the ageing process as measured with DNAm clocks. Research on the pathophysiology of AUD has revealed various effects of genes and their functional pathways on the development, maintenance and treatment of alcohol dependence.9 Changes in methylation state (known as epigenetic marks) are an important mechanistic link between specific genes and the behaviours that drive the course of the disease.7
The first evidence of an association between epigenetic ageing and AUD came from Rosen and colleagues10 who explored the effect of excessive alcohol consumption on age acceleration, including DNA methylation levels in whole blood and tissue samples from five independent cohorts of AUD subjects and controls (CON) using a chronological DNAm clock (Horvath clock11). Only two of the five datasets (blood and liver tissue) showed accelerated ageing, highlighting the need for further investigations on the mechanisms of the ageing process in AUD. A subsequent study from the same group demonstrated age acceleration by 2.2 years in AUD patients compared with CON using Levine's DNAm PhenoAge,12 a biological DNAm clock; the effect was greater in individuals with more severe AUD-associated phenotypes such as elevated liver transaminases and a higher number of heavy drinking days.
To this end, the present study examined the relationship between AUD and biological ageing in two independent cohorts. We first used the same basic methodologic approach as Luo et al.13 to analyse the discrepancy between true chronological age and biological age measured with Levine's DNAm PhenoAge, as well as chronological DNAm age measured with the Horvath clock and Zhang's age predictor as a next-generation statistical DNAm model. There have been no studies to date on the long-term effects of relapse and potential benefits of abstinence on biological ageing. The study by Luo et al. had a cross-sectional design,13 and the results did not specify whether differences in DNA methylation levels and epigenetic age acceleration are predisposing factors for addiction or if they are consequences of long-term alcohol use. To examine this interplay of genetic and environmental factors, we used methylation data from two time points (baseline and 12-month follow-up) previously published by our group9 to examine how alcohol exposure and epigenetic ageing interact and whether the absence of chronic, heavy alcohol consumption in abstinent AUD patients affects the deviation between true chronological age and biological age measured with Levine's DNAm PhenoAge.
The complex interplay between genetic and environmental factors poses an additional challenge to investigate epigenetic processes. Several genetic variants from recent Genome-Wide Association Studies (GWAS) have been associated with epigenetic age acceleration, for example TERT (the catalytic subunit of telomerase),14 DHX57 (an ATP-dependent RNA helicase) or MLST8 (a subunit of both mTORC1 and mTORC2 complexes),15 however, without reference to AUD. It is recognized that AUD is highly polygenic. We therefore aimed to investigate in an additional exploratory approach how a polygenic risk score (PRS) for alcohol dependence derived from a large genome-wide association study assessing alcohol dependence and problematic alcohol use (PAU)16 affects epigenetic age acceleration.
2 MATERIAL AND METHODS
2.1 Subjects
All subjects were recruited between 2012 and 2020 as part of a larger study (ClinicalTrials.gov identifiers: NCT01679145 and NCT02615977) investigating behavioural, genetic and neuroimaging alterations associated with reward-based learning in AUD over two funding periods. Thus, this study comprised two independent AUD cohorts from funding period 1 (DSCase@BaseVSLong) and funding period 2 (DSCaseVSCon). In addition, a healthy CON group was recruited in which subjects were matched with the AUD patients of DSCaseVSCon with respect to smoking status and several socio-economic variables (Data S1). Whole epigenome data for DSCase@BaseVSLong (changes in methylation levels over time) have been previously analysed and published by our group9 (Data S2A and S2B). Briefly, whole-genome methylation patterns of individual CpG sites over time did not differ between abstinent and relapsing patients. However, there was a negative association between global mean methylation at the 12-month follow-up and alcohol consumption within our sample. All patients fulfilled the diagnostic criteria for alcohol dependence according to International Classification of Disease-10 and Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) text revision17 for a minimum of 3 years. Patients with history of current or past substance use disorder (except alcohol and nicotine dependence), other major psychiatric disorders (as assessed using the computer-based Composite International Diagnostic Interview)18, 19 or neurologic disease were excluded. All subjects were free of psychotropic medication known to interact with the central nervous system for at least four half-lives (including illegal drugs and detoxification treatments as determined by a urine test). Patients were enrolled in the study shortly after detoxification (3–21 days) in both funding periods. Patients in DSCase@BaseVSLong were followed up for 12 months with the Alcohol Timeline Followback method.20 Relapse during this period was defined as consumption of 60 or 40 g of alcohol on any occasion for males and females, respectively, according to World Health Organization21 criteria of current high-risk versus low-risk consumption. Individual assessment included alcohol breath tests to validate self-reports. For DSCaseVSCon, methylation data were available from baseline and 2-week follow-up assessments only. Smoking status was assessed with the Fagerström Test for Cigarette Dependence.22 AUD patients and CON subjects were matched according to smoking status.
2.2 Genotype QC and imputation
The genotype QC and imputation was performed using Rapid Imputation for COnsortias PIpeLIne (RICOPILI) GWAS pipeline.23 The subjects and SNPs passed the QC if the following parameters were satisfied: SNP missingness < 0.05 (before sample removal); subject missingness < 0.02; autosomal heterozygosity deviation (|F het| < 0.2); SNP missingness < 0.02 (after sample removal); difference in SNP missingness between cases and controls < 0.02; and SNP Hardy–Weinberg equilibrium (p > 10e−6 in controls or p < 10e−10 in cases). Three population outliers were excluded by visually selecting a threshold from 2D plots of principal components 1 and 2 from a principal component analysis (PCA, see Figure S6). One subject failed the above missingsness filter, resulting in n = 185 subjects. For relatedness testing, 65 828 autosomal SNPs which were left after linkage disequilibrium (LD) pruning (r2 > 0.02) with minor allele frequency (MAF) > 0.05 were used. No pairs of subjects with an estimated proportion of IBD (PIHAT) > 0.2 were identified.
The genotype imputation was conducted using the pre-phasing/imputation stepwise approach implemented in Eagle (https://alkesgroup.broadinstitute.org/Eagle/)/MINIMAC3 (https://genome.sph.umich.edu/wiki/Minimac) with variable chunk size of 132 genomic chunks and default parameters. The imputation reference set consisted of 54 330 phased haplotypes with 36 678 882 variants from the publicly available Haplotype Reference Consortium (HRC) reference (https://ega-archive.org/datasets/EGAD00001002729).
2.3 Quantification of DNA methylation level and QC
Data preprocessing for both datasets was performed using the respective default functions of the The Chip Analysis Methylation (ChAMP) pipeline,24 mainly applying the default settings in the ChAMP functions. This includes the conversion of raw iDat files into beta values as well as the following six preprocessing steps that were performed as recommended by the authors of ChAMP: (1) Filter for probes with detection p value (default > 0.01). (2) Filter out probes with <3 beads in at least 5% of samples per probe. In this process, sample's failed probes' ratio above a threshold of 0.1 is regarded as failed measurements; in the DSCaseVSCon, one sample was excluded in this preprocessing step. (3) Filter out all non-CpG probes contained in the dataset. (4) Filter out all SNP-related probes. (4) Filter out all multi-hit probes. (5) Filter out all probes located in chromosome X and Y.
Based on our previous experience with batch effects that can lower data quality and yield false positives, we used 2-week follow-up measurements as systematic twofold data of all subjects in DSCaseVSCon with a stratified randomized distribution of the samples. All samples were analysed with the Illumina Infinum Human MethylationEPIC BeadChip (Illumina, San Diego, CA, USA).
As the 2-week age difference is negligible for the research question at hand and because there is no systematic difference in predicted age between the two measurements (tLevine[119] = 0.01, p = 0.99; Cohen's d = 0.00), the measurements were used as measurement repetitions and subsequently averaged for each subject in order to reduce measurement errors and to exclude systematic batch effects. One sample was excluded during QC due to showing a high fraction of failed probes. As there was no repeat measurement, the subject was completely excluded.
In order to meet the requirements for DNAm clock calculations, averaged samples were normalized for type I and II probe differences using the beta mixture quantile normalization (BMIQ) method.25 As previously reported,9 data for the first funding period (DSCase@BaseVSLong) had no measurement repetitions, and significant batch effects for sample plate, chip and row were corrected using ComBat from the sva package26 without target variables after normalizing the data by BMIQ as for DSCaseVSCon. Additionally, a significant batch effect between DSCase@BaseVSLong and DSCaseVSCon was adjusted with ComBat using only AUD patients from DSCaseVSCon as a reference group again without target variables.
2.4 Calculating DNAm age
We evaluated DNAm age using three different statistical models: the chronological Horvath clock comprising 344 CpG sites11; the biological Levine's DNAm PhenoAge DNAm clock model comprising 513 CpGs in 505 genes12; and Zhang's age predictor comprising a best linear unbiased prediction model based on 319 607 CpGs.27 While the Horvath clock was developed for Illumina 27K and 450K arrays and not for the EPIC arrays used in the present study, Levine's DNAm PhenoAge and Zhang's age predictor were developed for EPIC array data. CpG sites not measured due to the based array or missing due to exclusion were imputed for the Horvath clock and Levine's DNAm PhenoAge. While 27 (7.6%) CpG sites were missing from the Horvath clock specific instructions from the Horvath work group were applied, 12 (2.3%) missing CpG sites for Levine's DNAm PhenoAge were imputed using the mean methylation. Zhang's age predictor does not need imputation due to its high number of predictors. For the calculation of the Horvath clock and Levine's DNAm PhenoAge, the implementation in the Bioconductor package Methylclock28 was used; for Zhang's age, original code provided by the author was utilized. We then calculated the difference between actual age and age predicted with the DNAm clock models (Δ). Pearson's product–moment correlations between these values were positive, significant, and very large for all three DNAm clocks (Table 1), supporting the assumption of sufficient sample quality and correct sample preprocessing.
| DNAm clock | r DSCase@BaseVSLong | rDDSCaseVSCon |
|---|---|---|
| Horvath clock | 0.66** | 0.84** |
| Levines DNAm PhenoAge | 0.56** | 0.81** |
| Zhangs age predictor | 0.78** | 0.95** |
- Abbreviation: DNAm, DNA methylation.
- * p < 0.05.
- ** p < 0.01.
2.5 Calculating the PRS
For calculating the PRS, we used the summary statistics of the phenotypes AUD (cases/controls: 57 564/256 395) and PAU (n = 435 563) from a recent genome-wide association study from Zhou et al.16 Both the summary statistics were LD clumped (discarding variants within 500 kb of and in r2 ≥ 0.1 with another more significant marker), resulting in 120 446 (in AUD) and 235 621 (in PAU) SNPs, used for scoring. Specifically, we multiply the effect size from the training data set with the number of risk alleles on each SNP, summing up over each individual to have a whole-genome PRS in PLINK v1.90b4.1.29 A p value threshold of p = 1 representing the composite additive effect of all SNPs was applied, historically showing the strongest scoring results in psychiatric disorders.30 In Data S7, we additionally show results for more stringent p value thresholds (p = 0.05 and p = 0.001).
2.6 Single SNP analyses
For exploratory analyses only, we intended to assess the impact of ADH1B (rs1229984) and ALDH2 (rs671) on our epigenetic clocks. Both genetic variants have so far shown the strongest and most replicated association with alcohol dependence. However, due to the small sample size, the low frequency of risk alleles (MAF = 0.01) did not allow for further analyses. For replication purpose, we analysed the candidate SNP rs916264 in APOL2, previously shown to be associated with epigenetic age acceleration by Luo and colleagues.13
2.7 Statistical analysis
All statistical analyses were performed in the R (4.1.1) environment and Bioconductor (3.13) framework. To evaluate differences between AUD and CON groups in terms of the deviation between DNAm age determined with the Horvath clock, Levine's DNAm PhenoAge and Zhang's age predictor and true chronological age (Δ),27 we carried out analysis of variance (ANOVA) for the DSCaseVSCon dataset with group and age as independent and dependent variables, respectively. As in the study of Luo et al.,13 we added sex as a covariate and corrected for immune cell counts obtained using the ‘champ.refbase’ function of the ChAMP Package in Bioconductor.24 In a second step due to the high collinearity of the cell distribution, the variance inflation factors (VIFs) of the independent variables were calculated, and highly correlated predictors were removed accordingly.
To explore genes associated with CpGs of Levine's DNAm PhenoAge Δ, we calculated Pearson's product–moment correlations between each of the CpG sites included in the model and Δ. For this step, a false discovery rate (FDR) < 0.05 was defined as significant. We then calculated correlations between predicted DNAm PhenoAge and liver function indices (gamma-glutamyltransferase [γ-GT], aspartate aminotransferase [AST], alanine aminotransferase [ALT]). We also revisited previously published data of our DSCase@BaseVSLong cohort9 to investigate the possible development of/intergroup differences in Levine's DNAm PhenoAge over a 12-month period (which had not been previously examined) with a mixed linear model to predict Levine's clock Δ, with relapsers/abstainers, sex and immune cell counts as covariates over time. Again, VIF was checked for high collinearity and collinear predictors were removed accordingly.
As for the analyses of DSCaseVSCon, we calculated Pearson's product–moment correlations between each of the CpG sites included in the Levine's DNAm PhenoAge model and Δ, with FDR < 0.05 defined as significant.
Association between the PRS (see methods above) and phenotypes (AUD and PAU) was tested using logistic regression and adjusted for population stratification (using PC's as covariates 1–5) in DSCaseVSCon (n = 116, 85 cases/31controls, DSCase@BaseVSLong was excluded due to missing controls). The explained variance was estimated with Nagelkerke's R2 (NKr2) by comparing scores generated from a full model (containing covariates and PRS) and a reduced model (covariates only).
Variance in Levine's DNAm PhenoAge Δ explained by PRS for AUD and PAU was then tested using linear regression analysis in DSCaseVSCon and DSCase@BaseVSLong (n = 185). Similar to previous analysis, the analysis was also corrected for population stratification using PCs (1 to 5). The beta coefficients and adjusted R2 were estimated.
Association between the SNP rs916264 in APOL2 and Levine's DNAm PhenoAge Δ was tested using PLINK (v1.90b4.1) based linear regression analysis. The analysis was adjusted for population stratification using PCs (1 to 5) in DSCaseVSCon and DSCase@BaseVSLong (n = 171 subjects, 14 subjects had to be excluded due to missing genotype, MAF = 0.19).
3 RESULTS
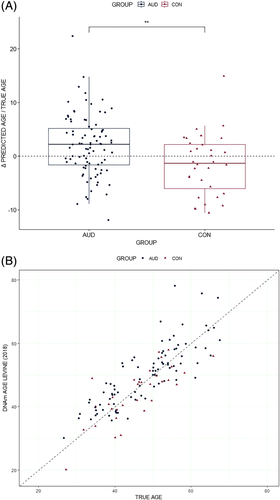
ANOVA for differences between AUD and CON groups in the DSCaseVSCon with regard to age Δ did not show a significant effect for Horvath's DNAm clock (F(1, 112) = 0.43, p = 0.51; η2 [partial] = 0.00) (Data S3A). The analysis for Zhang's age predictor yielded comparable results (F[1, 112] = 0.40, p = 0.53; η2 [partial] = 0.00) (Data S3B). However, Levine's DNAm PhenoAge showed a significant medium size effect (F[1, 112] = 11.10, p = 0.00; η2 [partial] = 0.09) (Figure 1A,B and Table 2). AUD patients showed a mean (±SD) accelerated ageing of 2.94 (±5.69) years compared with their chronological age whereas in CON subjects, and the mean (±SD) Δ was −0.66 (5.7) years, yielding a mean difference of 3.6 years between the two groups.
| Parameter | Sum of squares | df | Mean square | F | p | Partial η2 | VIF |
|---|---|---|---|---|---|---|---|
| AUD/CON | 304.76 | 1 | 304.76 | 11.10 | 0.00 | 0.09 | 1.05 |
| Sex | 0.97 | 1 | 0.97 | 0.04 | 0.85 | 0.00 | 1.15 |
| CD8T | 49.28 | 1 | 49.28 | 1.79 | 0.18 | 0.02 | 1.09 |
| CD4T | 586.62 | 1 | 586.62 | 21.36 | 0.00 | 0.16 | 1.21 |
| NK cells | 20.55 | 1 | 20.55 | 0.75 | 0.39 | 0.01 | 1.13 |
| B cells | 35.18 | 1 | 35.18 | 1.28 | 0.26 | 0.01 | 1.19 |
| Monocytes | 55.78 | 1 | 55.78 | 2.03 | 0.16 | 0.02 | 1.27 |
| Residuals | 3075.67 | 112.00 | 27.46 |
- Note: Granulocytes were removed as predictor due to excessive collinearity with the other predictors. Variance inflation factors (VIFs) of the remaining predictors <1.5.
- Abbreviations: AUD, alcohol use disorder patients; B cells, B lymphocytes; CD8T, CD8+ cytotoxic T lymphocytes; CD4T, CD4+ cytotoxic T lymphocytes; CON, control subjects; DNAm, DNA methylation; NK, natural killer.
Pearson's product–moment correlations in the DSCaseVSCon revealed four CpG sites with an FDR-corrected significant influence of Δ (Data S4): cg05851163 located in the 5′ untranslated region of the dermatan sulphate epimerase-like (DSEL) gene (r = 0.36, 95% confidence interval [CI]: 0.20–0.51, t[118] = 4.25, pFDR < 0.05); cg23668631 located in the TSS1500 shore of the calcium/calmodulin dependent protein kinase kinase 1 (CAMKK1) gene (r = −0.35, 95% CI: −0.50 to −0.18, t[118] = −4.03, pFDR < 0.05); cg18468844 located in the TSS1500 OpenSea of the platelet-activating factor receptor (PTAFR) gene (r = −0.35, 95% CI: −0.49 to −0.18, t[118] = −4.00, pFDR < 0.05); and cg01211097 located in the TSS1500 shore of the ubiquitin-specific peptidase 10 (USP10) gene (r = −0.34, 95% CI: −0.49 to −0.17, t[118] = −3.87, p < 0.05). On the other hand, Pearson's product–moment correlations between predicted Levine's DNAm PhenoAge and liver function indices were nonsignificant (γ-GT: r = −0.01, t[114] = −0.06, p = 0.95; AST: r = 0.07, t[114] = 0.70, p = 0.49; and ALT: r = −0.04, t[114] = −0.41, p = 0.68).
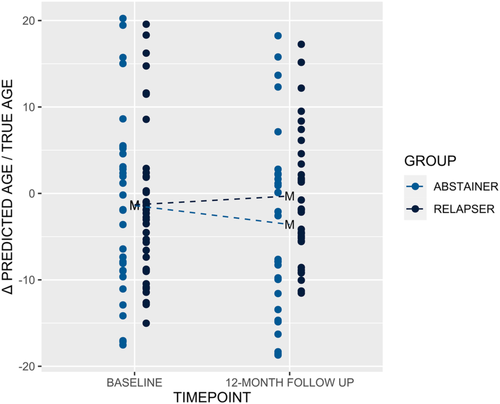
The mixed linear model analysis of DSCase@BaseVSLong to predict the change in Levine's DNAm PhenoAge Δ over time showed a high total explanatory power (conditional R2 = 0.86) with a significant interaction effect between time and group (β = 3.65, t[126] = 2.98, p < 0.01). While relapsers exhibited accelerated ageing by a mean (±SD) of 2.69 (±7.49) years during the 12-month period, abstainers were by a mean (±SD) of 4.79 (7.72) years below their expected chronological age (Figure 2 and Table 3). Pearson's product–moment correlation analysis of DSCase@BaseVSLong revealed only one CpG site—cg09809672 located in the TSS1500 shore of the ectodysplasin; a receptor associated death domain (EDARADD) gene—that was significantly associated with Δ after FDR correction (r = 0.35, 95% CI: 0.19 to 0.49, t[130] = 4.24, pFDR < 0.05) (Data S5).
| Parameter | Coefficient | 95% CI | t | df | p | Std. coef. | Fit | VIF | |
|---|---|---|---|---|---|---|---|---|---|
| (Intercept) | 15.66 | 7.17 | 24.16 | 3.65 | 126 | 0.00 | 0.03 | ||
| Group | −0.53 | −4.39 | 3.33 | −0.27 | 126 | 0.79 | −0.06 | 1.13 | |
| Time | −1.92 | −3.69 | −0.15 | −2.15 | 126 | 0.03 | −0.22 | 2.49 | |
| Sex | −6.59 | −11.79 | −1.40 | −2.51 | 126 | 0.01 | −0.27 | 1.02 | |
| CD8T | −12.42 | −44.19 | 19.34 | −0.77 | 126 | 0.44 | −0.04 | 1.89 | |
| CD4T | −14.56 | −43.54 | 14.42 | −0.99 | 126 | 0.32 | −0.07 | 3.71 | |
| NK cells | −21.02 | −47.67 | 5.63 | −1.56 | 126 | 0.12 | −0.06 | 1.17 | |
| B cells | −138.09 | −189.70 | −86.48 | −5.30 | 126 | 0.00 | −0.28 | 2.26 | |
| Monocytes | −9.80 | −43.01 | 23.41 | −0.58 | 126 | 0.56 | −0.03 | 1.62 | |
| Group × time | 3.65 | 1.23 | 6.07 | 2.98 | 126 | 0.00 | 0.43 | 2.69 | |
| R2 (conditional) | 0.86 | ||||||||
| R2 (marginal) | 0.21 | ||||||||
- Note: Granulocytes were removed as predictor due to excessive collinearity with the other predictors.
- Abbreviations: B cells, B lymphocytes; CD8T, CD8+ cytotoxic T lymphocytes; CD4T, CD4+ cytotoxic T lymphocytes; CI, confidence interval; DNAm, DNA methylation; NK, natural killer; Std. coef., standardized coefficient; VIF, variance inflation factor of the remaining predictors <1.5.
The calculated PRS did show a significant association with alcohol dependence at p-value threshold p = 1.0 for both AUD (NKr2 = 0.0497, p = 0.0457) and PAU (NKr2 = 0.1138, p = 0.0022). For p-value thresholds p = 0.05 and p = 0.001, see Data S7.
There was no significant association between PRS and Levine's DNAm PhenoAge Δ. There was also no significant association between the SNP rs916264 in APOL2 and Levine's DNAm PhenoAge Δ.
4 DISCUSSION
Only two studies to date have investigated epigenetic age acceleration in patients with AUD.10, 13 Our results confirm the earlier finding that AUD is associated with accelerated ageing, albeit to an even greater extent than previously reported (4.7 vs. 2.2 years). However, we are the first to report the potentially recovering effects of long-term abstinence from alcohol and the potential influence of polygenic risk on epigenetic ageing.
In this study, we investigated epigenetic ageing in two patient cohorts. Results from DSCaseVSCon supported those of Luo et al.,13 revealing a difference between AUD patients and CON subjects in terms of the discrepancy between epigenetic age and true chronological age. In line with the previous findings, differences in age acceleration were detected with Levine's DNAm PhenoAge only and not with chronological DNAm clocks (Horvath clock and Zhang's age predictor). This is expected given the variations in methodologic approaches for the two types of DNAm clocks. Chronological clocks are designed to predict true chronological age in very large heterogeneous samples5; possible biological markers are not included, as these large training samples include people with preexisting conditions, and therefore, only CpG sites that change uniformly over time independent of environmental influences are considered. Also, Zhang estimate is tailored to estimate chronological age as precisely as possible and explicitly not to be affected by mortality nor cell composition in ideal case. In contrast, biological DNAm clocks (including Levine's clock) favour the inclusion of CpGs that are altered in response to environmental factors. Thus, our results support the previous assertion13 that significant results can only be expected for DNAm clocks measuring biological age. Contrary to the results of Luo et al., we found no association between liver transaminases and predicted Levine DNAm PhenoAge, which underscores the fact that the molecular mechanisms of epigenetic clocks are complex and poorly understood.
Our second cohort of relapsing and abstaining patients (DSCase@BaseVSLong) yielded conflicting findings. The difference in predicted age Δ of 7.5 years between relapsers and abstainers from baseline to the 12-month follow-up was highly significant, indicating that abstinence from alcohol has beneficial effects on biological ageing in AUD patients. However, our results did not show an absolute positive age difference between predicted and actual age at baseline; an absolute age deviation only became apparent for relapsing patients at the 12-month follow-up. A possible reason for these unexpected results is the lower quality of measurements in the DSCase@BaseVSLong sample compared with the repeated measurements for DSCaseVSCon, which allowed necessary corrections and is also reflected in the lower correlations between the Horvath clock or Zhang's age predictor and true chronological age (Table 1). Given this limitation of our study, replication of our results is necessary. Another limitation is that in DSCaseVSCon and at baseline in DSCase@BaseVSLong, all AUD patients had been detoxified from alcohol for at least 3 but up to 21 days before measurements. Moreover, all patients had abstained from alcohol for at least 30 days until the first relapse and had had AUD (according to DSM-IV criteria) for a minimum of 3 years before enrollment. Given the clinical course of abstainers in DSCase@BaseVSLong over the 12-month period, we suspect that the actual epigenetic age difference between actively drinking AUD patients and CON subjects is even greater than what was shown by our analysis. It would therefore be highly valuable to repeat these experiments with active drinkers prior to their detoxification.
Previous studies have reported associations between epigenetic ageing and leading causes of death and disease burden.31 The consumption of alcohol is linked to epigenetic changes that may contribute to these long-term consequences.32, 33 Several candidate genes associated with AUD were identified in a large-scale epigenome-wide study using a cross-tissue/cross-phenotype approach.34 Our exploratory investigation of the (epi)genetic background of age deviations in AUD with a series of correlations for every individual CpG in the Levine DNAm PhenoAge model identified significant and large effects for CpG sites associated with four genes—namely, DSEL, CAMKK1, PTAFR and USP10.
Polygenic risk is a potential indicator of genetic liability in AUD; thus, we intended to explore its effect on the epigenetic ageing process. Both PRS scores based on Zhou et al.16 showed a significant association with alcohol dependence. However, we did not find an effect of the PRS on age acceleration, an exploratory single SNP analyses of ADH1B (rs1229984) and ALDH2 (rs671) on our epigenetic clocks failed due to the low number of risk alleles. We could also not replicate the previously reported association of epigenetic ageing with APOL2 (rs916264).13 With respect to sample size and construction of the Levine DNAm PhenoAge,12 which has been designed with a certain robustness against genetic influences, a zero-finding is comprehensible. However, with respect to the fact that the interplay between genetic variables and epigenetic ageing remains poorly understood,35 we suggest to apply the same approach in a bigger sample.
In the DSCase@BaseVSLong, we identified one CpG associated with accelerated epigenetic ageing in relapsers compared with abstainers located in the shore of the EDARADD gene. As we limited the correlation analyses to differences in age acceleration detected using Levine clock CpG sites, it is not surprising that the genes with the highest correlations are related to metabolic functions and cell regeneration. While these analyses are limited by the number of CpGs included in the Levine DNAm PhenoAge model, the significant CpG sites or their associated genes are of interest for subsequent investigations on the (epi)genetic background of accelerated ageing in AUD. Given the limited size of our two cohorts, our results should be considered as preliminary. In the genome-wide association study carried out by Luo et al., a single nucleotide polymorphism in the apolipoprotein L2 (APOL2) gene was implicated in accelerated epigenetic ageing.13 However, the study had a cross-sectional design and did not consider the effects of long-term abstinence and relapse. On the other hand, tissue-specific associations between alcohol dependence and epigenetic age were observed with the Horvath and Hannum clocks,10 although these are considered less effective in reflecting environmental influences on the ageing process.
In summary, DNAm clocks serve as valuable biomarkers of biological ageing. It will be interesting to define subgroups of AUD patients that are more prone to epigenetic age acceleration or more responsive to the positive effects of abstinence. Genetic determinants are one of many possible confounders for associations with changes in transaminases and other tissue-specific effects. Subsequently, the complex interplay between genetic and environmental factors poses an additional challenge to interpret the data and determine causal relationships.
Epigenetic measures of ageing have potential utility in clinical settings as a complement to gold-standard methods for disease assessment and management.31 For example, our results may be used in clinical practice to motivate patients with AUD to take the difficult path of long-term sobriety and associated alcohol withdrawal, as it can lead to measurable biological recovery. Finally, DNAm clocks may provide novel targets for pharmaceutical interventions, as demonstrated in other disease such as bipolar disorder36 and schizophrenia.37
ACKNOWLEDGEMENTS
We thank the LeAD study teams in Dresden and Berlin for assistance with data acquisition, especially Lena Elisabeth Fliedner and Robin Frank, Vanessa Buchholz and Tanja Wesse for the bisulfite conversion experiments. We thank Prof. Joel Gelernter, Prof. Hang Zhou and the VA Million Veteran Program (MVP) for sharing their data with us. Open Access funding enabled and organized by Projekt DEAL.
CONFLICT OF INTEREST
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ETHICS APPROVAL
Ethics approval for the study was obtained from the ethics committee of Charité-Universitätsmedizin Berlin (EA1/157/11) and Universitätsklinikum Dresden (EK228072012). Participants received a monetary compensation of 10 €/hour for study participation.
AUTHOR CONTRIBUTIONS
TZ and EF conceived the study; TZ, EF, HF, LF, IV, AN and HW drafted the manuscript; TZ, EF, HF, LF, IV, AN, SA, SR and HW analysed and interpreted the data; SA und SR calculated the PRS, and TZ created the figures; HF supervised the study; and TZ, EF, HF, LF, IV, AN, SA, SR and HW revised the manuscript for important intellectual content. All authors contributed to manuscript revision, and read and approved the submitted version.
Open Research
DATA AVAILABILITY STATEMENT
This manuscript contains previously unpublished data. The name of the repository and accession number(s) are not available due to patient confidentiality and participant privacy. Methylation array data can simultaneously identify individuals and convey protected health information 43434343434343. Patients did not provide written, informed consent for the publication of individual methylation profiles. Requests to access the datasets should be directed to Eva Friedel ([email protected]).




