Overexpression of eukaryotic translation initiation factor 4E-binding protein 1 induces the alteration of immune status in H1299 lung cancer cells
Abstract
Background
Eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) is an important factor regulating protein translation. It also impacts proliferation, apoptosis, invasion, and the cell cycle of cancer cells. The aim of this study was to investigate the relationship between 4E-BP1 and human immune status, recognizing immunomodulatory molecules involved in the overexpression of 4E-BP1.
Methods
A lentivirus expression system was used to overexpress 4E-BP1 in the H1299 cell line. Western blot was performed to investigate the expression level of 4E-BP1 and P-4E-BP1, and quantitative polymerase chain reaction was used to quantify gene expression of immunomodulatory molecules.
Results
The expression level of 4E-BP1 increased significantly after lentivirus infection (P < 0.05). Overexpression of 4E-BP1 upregulated the expression of interleukin (IL)-1β (P < 0.05), IL-5 (P < 0.001), IL-23 (P < 0.001), macrophage inflammatory protein-1β (P < 0.001), Eota-3 (P < 0.05), and MCP-4 (P < 0.05). Most of the increases were observed at the seventh day. The variation trend of IL-10, cell division cycle protein 2, proliferating cell nuclear antigen, and phosphatase and tensin homolog was not clear.
Conclusion
Overexpression of 4E-BP1 altered immune status by upregulating the expression of a series of immunomodulatory molecules, indicating that 4E-BP1 could serve as a potential therapeutic target against cancer.
Introduction
Lung cancer is the leading cause of cancer related death all over the world. Non-small cell lung cancer (NSCLC) is the most common type, accounting for nearly 85% of all primary lung carcinomas.1 During the past few years, some proteins, such as protein kinase B (Akt),2 mammalian target of rapamycin (mTOR),3 epidermal growth factor receptor (EGFR),4 and V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) were reported to play an essential role in tumorigenesis.5 However, the specific regulatory mechanisms are still undefined.
The most common type of eukaryotic translation initiation factor 4E binding protein (4E-BP) family, 4E-BP1, lies downstream of the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/Akt/mTOR signaling pathway. It is a crucial factor controlling protein synthesis, cell survival, and proliferation.6 By binding to eukaryotic translation initiation factor 4E (eIF4E), 4E-BP1 can prevent its association with ribosome, which leads to the inhibition of protein translation. Mammalian target of rapamycin complex 1 (mTORC1), one of the distinct multiprotein complexes mTOR, is an important upstream protein and also a serine/threonine kinase, which regulates 4E-BP1 directly. In cancer cells, with an increased level of phosphorylated mTORC1, 4E-BP1 is abundantly phosphorylated. Phosphorylated 4E-BP1 (p-4E-BP1) has a decreased affinity with eIF4E, so dissociates from it. Released eIF4E then binds to messenger ribonucleic acid (mRNA) cap structure, initiating protein translation in eukaryote.7 When stimulated by serum, growth factor, and hormones, the level of p-4E-BP1 also increases.8 Previous studies have reported that the overexpression of 4E-BPs can strongly inhibit the tumorigenic properties of cancer cells.9
The immune status of patients with cancer is also altered. A retrospective study of metastatic cancers showed that a high expression of CXCR4/CXCL12 was associated with more advanced stages and poorer prognosis.10 However, the relationship between immunomodulatory molecules, such as tumor necrosis factor, interferon, interleukin (IL)-6, and signaling pathways remains ambiguous.
Here, via reverse transcription and quantification polymerase chain reaction (qPCR), we examined which types of immunomodulatory molecules were involved in the overexpression of 4E-BP1 and the promotion of inflammatory status of H1299 lung cancer cell.
Methods
Lentiviral expression system for eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) overexpression
Human lung NSCLC cell line H1299 was purchased from American Type Culture Collection (ATCC, Manassas, VA USA) and was cultured in PRMI1640 supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine, at 37°C in a 5% CO2 humidified incubator.
The lentiviral expression system for 4E-BP1 overexpression was constructed by NeuronBiotech Corp (Shanghai, China). A sequence expressing 4E-BP1 (forward primer: 5′-CCTTTCCGGGACTTTCGCTTT-3′; reverse primer: 5′-GCAGAATCCAGGTGGCAACA-3′; probe: 5′-FAM-ACTCATCGCCGCCTGCCTTGCC-TAMRA-3′) was cloned into a lentivirus transfection vector (pLOV-CMV-eYFP-4EBP1-EF1a-PuroR). An empty vector was then constructed in the same manner, as a negative control (pLOV-CMV-eYFP-EF1a-PuroR). Green fluorescent protein (GFP) was used as a marker to sort transfected H1299 cells, using flow cytometry. Cells infected with lentivirus expressing 4E-BP1 were named as the overexpression (OE) group and those with a negative control sequence were named the negative control (NC) group. Cells that were not infected by lentivirus were named the H1299 group.
Reverse transcription and quantitative real time polymerase chain reaction (qPCR)
After resuscitation, the H1299, OE, and NC groups were cultured according to ATCC protocol for several days. Total RNA were then isolated using an RNeasy mini kit (Qiagen, Dusseldorf, Germany), from each group on the first, third, fifth, and seventh days, separately. A ReverTra Ace qPCR kit (FSQ −101; Toyobo, Kagoshima, Japan) was used to synthesize cDNA. The conditions for reverse transcription were 65°C for five minutes, 37°C for 15 minutes, then 98°C for five minutes.
All of the PCR primers used were designed according to data provided by GenBank (Table 1). RealMaster Mix (SYBR Green; FP202; Tiangen, Beijing, China) was used to perform qPCR. All procedures were fulfilled in an iCycler iQTM Optical Module (Beckman Coulter, Fullerton, CA, USA) under the following conditions: initial denaturation at 95°C for 30 seconds, followed by 40 cycles at 95°C for 30 seconds, then 58°C for 30 seconds and 72°C for 30 seconds, followed by a melt curve from 55 to 95°C in 0.5°C increments and 10 second intervals. All tests were repeated three times.
| Gene | Forward primer | Reverse primer | GenBank number |
|---|---|---|---|
| Interleukin | |||
| IL-1β | ACGAATCTCCGACCACCACT | CCATGGCCACAACAACTGAC | M15330 |
| IL-5 | CAGGAGACTGAGAGGTCATGC | CCGCAGCGCATAAAGAGT | NM_005732 |
| IL-8 | CTGGCCGTGGCTCTCTTG | CCTTGGCAAAACTGCACCTT | NM_000584 |
| IL-9 | AGCCCCAGGCAGCTACAC | AAGAGTCCCGCCACATCAC | NM_000590 |
| IL-10 | GCTGGCTTCCTTTCTCTGAA | CCGCAGCGCATAAAGAGT | NM_000572 |
| IL-23 | GATGCAGGGAGGGGAATC | CCGCAGCGCATAAAGAGT | NM_031252 |
| Chemokine | |||
| Eota-3 | TGTACCAAGTCCCTCCTACTAGC | TCATGTTGGAGGCTGAAGGT | NM_001256774 |
| MCP-1 | TTCGGTCTTGGGAAATGACT | CCGCAGCGCATAAAGAGT | NM_001028468 |
| MCP-2 | GTCACCAACCGCCAAGAG | CCGCAGCGCATAAAGAGT | XM_008274177 |
| MCP-4 | CAGCTCAGCAGATTCAGGAT | ACCTGGCTGAGCAAGTCC | NM_001021691 |
| MIP-1β | CTGCTCTCCAGCGCTCTCA | GTAAGAAAAGCAGCAGGCGG | NM_002984 |
| RANTES | GACACCACACCCTGCTGCT | TACTCCTTGATGTGGGCACG | NM_002985 |
| Tumor-related genes | |||
| cdc2 | AGCCCCAGGCAGCTACAC | CCGCAGCGCATAAAGAGT | NM_057449 |
| PCNA | CAGCCAATGAGGGCTAGG | AGGTGTTGGAGGCACTCAAG | BC000491 |
| Smad-3 | ACGTCTCCCATCCTCCAAG | GAAAAGATGCACTTTGCTTTAATA | AC012568 |
| PTEN | ACCATAACCCACCACAGC | CAGTTCGTCCCTTTCCAG | NM_058074 |
| GAPDH | GAAGGTGAAGGTCGGAGTC | GAAGATGGTGATGGGATTTC | J04038 |
- cdc2, cell division cycle protein 2; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IL, interleukin; MCP, monocyte chemotactic protein 1; MIP-1β, macrophage inflammatory protein-1β; PCNA, proliferating cell nuclear antigen; PTEN, phosphatase and tensin homolog; qPCR, quantitative polymerase chain reaction; RANTES, regulated upon activation, normal T cell expressed and secreted; Smad-3, Sma and Drosophila Mad protein 3.
Western blot analysis
Total proteins of the H1299, NC, and OE groups were extracted using a protein extraction kit (KeyGEN, Nanjing, China). Lysates were centrifuged with 14 000 rpm for 15 minutes at 4°C. Protein concentrations were measured with a BCA Protein Assay reagent (Thermo scientific, Rockford, IL, USA). For 4E-BP1, 7.5 ug total protein was loaded, with 15 ug for P-4E-BP1; 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel was used to separate different proteins and then transferred to 0.2 um polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). Membranes were blocked with 5% skimmed milk in tris buffered saline with 0.1% Tween-20 (TBST) for one hour at room temperature, then incubated with primary antibody (Table 2), at 4°C overnight. The following day, membranes were washed with TBST three times for 10 minutes each time, then incubated with horseradish peroxidase-conjugated antirabbit secondary antibody for one hour at room temperature. After washing a further three times at eight minutes each time, target proteins were detected with X-ray film after exposure to enhanced chemiluminescence reagent (Amersham Biosciences, Fairfield, CT, USA).
| Name | Company | Cat. number | Molecular weight (kDa) | Dilution |
|---|---|---|---|---|
| β-actin | CST | 8457 | 45 | 1:3000 |
| 4E-BP1 | CST | 9644 | 15–20 | 1:1000 |
| P-4E-BP1 | CST | 9459 | 15–20 | 1:1000 |
| Anti-rabbit antibody | CST | 7074 | — | 1:2000 |
- 4E-BP1, eukaryotic translation initiation factor 4E binding proteins; P-4E-BP1, phosphorylated 4E-BP1.
Statistical analysis
Bio-Rad iQ5 software ((Bio-Rad Laboratories Inc., Hercules, CA, USA) was used to analyze qPCR data, and ImageJ software (National Institutes of Health, Bethesda, MD, USA) was used to conduct the gray analysis of Western blot. All figures were created by GraphPad Prism 6.0 (GraphPad Software, Inc., San Diego, CA, USA), while all calculations were completed using SPSS 17.0 for Windows (SPSS Inc., Chicago, IL, USA). In the present study, glyceraldehyde 3-phosphate dehydrogenase was selected as an internal control, and the NC group on the first day were used as a negative control. Data were expressed as mean ± standard error of the mean (SEM). Results of the t test indicated a significant difference compared to the control group, if P < 0.05.
Results
Detection of 4E-BP1 and P-4E-BP1 expression level by Western blot
The expression levels of 4E-BP1 and P-4E-BP1 were tested in the H1299, NC, and OE groups by Western blot. As shown in Figure 1, the expression level of 4E-BP1 was significantly increased in the OE group (P < 0.05), indicating that the lentivirus expression system was effective. However, the expression level of 4E-BP1 was also increased in the NC group (P < 0.05) compared with the H1299 group. For P-4E-BP1, the expression level did not alter significantly between the different groups (P > 0.05).
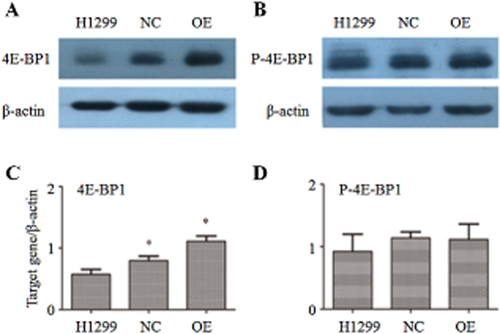
Analysis of eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) and P-4E-BP1 expression level by Western blot. (a) Western blot result of 4E-BP1; (b) Western blot result of P-4E-BP1; (c) gray analysis of 4E-BP1; (d) gray analysis of P-4E-BP1. *P < 0.05.
Quantification survey of interleukins by qPCR
Six interleukins were analyzed in this study. Their expression levels were tested by qPCR in order to recognize relevant genes in response to 4E-BP1 overexpression in the H1299 cell line. Different time points were also tested in order to identify the time effect of 4E-BP1 overexpression on immune status.
As shown in Figure 2, the expression levels of the six interleukins altered greatly between the different groups and time points. The most significant increase was found on the seventh day in the OE group for IL-1β (P < 0.05), IL-5 (P < 0.001), and IL-23 (P < 0.001). For IL-8 (P < 0.05) and IL-9 (P < 0.05), the expression level increased slightly with time, but the difference between the groups was unclear. Expression of IL-10 was rare in the H1299 cell line; however, there was a significant increase of IL-10 expression in the OE group on the first (P < 0.05) and seventh days (P < 0.001).
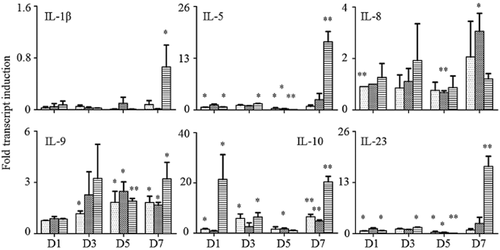
Detection of interleukin expression variation by quantitative polymerase chain reaction. D1, ribonucleic acid (RNA) extracted on the first day; D3, RNA extracted on the third day; D5, RNA extracted on the fifth day; D7, RNA extracted on the seventh day. *P < 0.05, **P < 0.001.  , H1299;
, H1299;  , negative control (NC);
, negative control (NC);  , overexpression (OE). IL, interleukin.
, overexpression (OE). IL, interleukin.
Quantification survey of chemokines by qPCR
Six chemokines were examined in the present study. The results of qPCR are exhibited in Figure 3. Overexpression of 4E-BP1 could greatly increase the expression levels of macrophage inflammatory protein (MIP)-1β (P < 0.001), Eota-3 (P < 0.05), and monocyte chemoattractant protein (MCP)-4 (P < 0.05) on the seventh day, as well as regulated on activation, normal T cell expressed and secreted (RANTES) (P < 0.05) on the third day. For MCP-2, the highest expression level was found on the first day in the OE group, and then decreased with time in all groups. However, a slightly increased expression was found again on the seventh day (P < 0.05). There was no significant difference in MCP-1 expression between the different groups and time points.
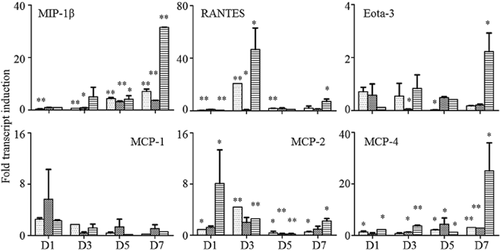
Detection of chemokine expression variation by quantitative polymerase chain reaction. D1, ribonucleic acid (RNA) extracted on the first day; D3, RNA extracted on the third day; D5, RNA extracted on the fifth day; D7, RNA on the seventh day. *P < 0.05, **P < 0.001.  , H1299;
, H1299;  , negative control (NC);
, negative control (NC);  , overexpression (OE). MCP, monocyte chemotactic protein 1, MIP-1β, macrophage inflammatory protein-1β; RANTES, regulated upon activation, normal T cell expressed and secreted.
, overexpression (OE). MCP, monocyte chemotactic protein 1, MIP-1β, macrophage inflammatory protein-1β; RANTES, regulated upon activation, normal T cell expressed and secreted.
Quantification survey of tumor-related genes by qPCR
Four tumor-related genes were tested by qPCR: phosphatase and tensin homolog (PTEN), a well-known tumor suppressor; proliferating cell nuclear antigen (PCNA), a molecular marker for proliferation; Sma and Drosophila Mad protein 3 (Smad-3), an important transcriptional factor; and cell division cycle protein 2 (cdc2), a regulator of cell cycle. Results are shown in Figure 4. The expression level of Smad-3 increased with time in all groups (P < 0.05). However, no significant difference was found when 4E-BP1 was overexpressed. For cdc2, PCNA, and PTEN, expression levels were low in all groups. Meanwhile, no significant changes were found between the groups and time points.
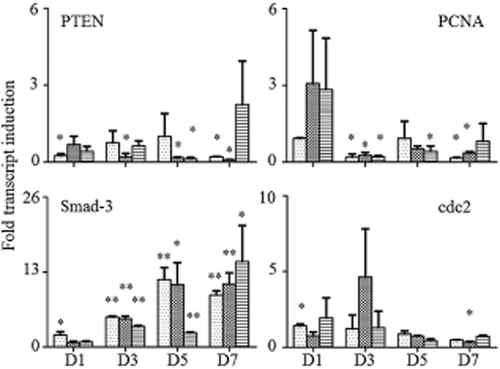
Detection of tumor-related gene expression variation by quantitative polymerase chain reaction. D1, ribonucleic acid (RNA) extracted on the first day; D3, RNA extracted on the third day; D5, RNA extracted on the fifth day; D7, RNA extracted on the seventh day. *P < 0.05, **P < 0.001.  , H1299;
, H1299;  , negative control (NC);
, negative control (NC);  , overexpression (OE). cdc2, cell division cycle protein 2; PCNA, proliferating cell nuclear antigen; PTEN, phosphatase and tensin homolog; Smad-3, Sma and Drosophila Mad protein 3.
, overexpression (OE). cdc2, cell division cycle protein 2; PCNA, proliferating cell nuclear antigen; PTEN, phosphatase and tensin homolog; Smad-3, Sma and Drosophila Mad protein 3.
Discussion
4E-BP1 plays an important role in tumorgenesis. By interfering in the formation of the eukaryotic translation initiation factor, it regulates protein translation, and then impacts cell proliferation, cell cycle invasion, and apoptosis;11 P-4E-BP1 is the functional form. In tumor cells, the upstream proteins Akt and mTOR largely phosphorylate 4E-BP1. The increase of P-4E-BP1 leads to the initiation of protein translation. As a result, tumorigenic properties of cells are inhanced.12, 13 In recent years, the relationship between 4E-BP1/P-4E-BP1 and prognosis has been extensively studied. Previous studies have reported that high expression levels of 4E-BP1 and P-4E-BP1 correlate strongly with poor prognosis in many cancers, such as breast,14 prostate, and colon cancers, as well as lung cancer.15, 16 However, few studies have investigated the correlation between 4E-BP1 and human immune status.
The current study examined the expressional variations of immunomodulatory factors in the H1299 cell line when 4E-BP1 was overexpressed. 4E-BP1 overexpression could increase the expression of numerous immunomodulatory factors, including interleukins (IL-1β, IL-5, IL-23) and chemokines (MIP-1β, Eota-3 and MCP-4). None of the factors we examined showed decreased expression. Our results indicated that 4E-BP1 could impact human immune status by upregulating the expression of immunomodulatory factors. However, most of the increased expression we observed occurred on the seventh day. A possible explanation for this phenomenon is that phosphorylation of 4E-BP1 is a delayed procedure; P-4E-BP1 is the functional form. When 4E-BP1 expression increases, it takes time to upregulate its phosphorylation level, and then subsequently impact the tumorigenic property of cells. This hypothesis could also explain Western blot results, which indicated that the P-4E-BP1 expression level was not altered when 4E-BP1 was overexpressed.
The key advantage of the current study was that a series of immunomodulatory factors were examined. Previous studies have not focused attention on the relationship between 4E-BP1 and immunomodulatory factors; only a few factors have been examined, neglecting the relationship between 4E-BP1 and immune status, as well as relationships among different immunomodulatory factors. Moreover, different time points were also examined in the present study. Our results could provide insight into the time effect of 4E-BP1 phosphorylation.
An essential limitation of the study was that only one cell line was detected. Comprehensive studies testing more cell lines, such as A549, H460, SK-MES, as well as HBE, are needed to explore the relationship between 4E-BP1 and the human immune system. The expression level of 4E-BP1 and P-4E-BP1 should also be tested at different time points by Western blot, which would help to verify the time effect of 4E-BP1 phosphorylation.
Conclusion
In conclusion, our study demonstrated that 4E-BP1 overexpression could increase the expression of a series of immunomodulatory factors. It suggests that 4E-BP1 could impact human immune status, and should be considered as a future therapeutic target against cancer.
Acknowledgment
This work was supported by grants from the National High Technology Research and Development Program of China (863 Program, no. 2014AA022202), the National Natural Science Foundation of China (81201851, 81241068, 81372504) the Chinese Postdoctoral Science Foundation (2013 M542281), and the Application of Infrastructure Program from the Department of Science and Technology, Sichuan Province, China (2013JY0012).
Disclosure
No authors report any conflict of interest.




