Journeying towards best practice data management in biodiversity genomics
Natalie J. Forsdick and Jana Wold are co-first authors.
Abstract
Advances in sequencing technologies and declining costs are increasing the accessibility of large-scale biodiversity genomic datasets. To maximize the impact of these data, a careful, considered approach to data management is essential. However, challenges associated with the management of such datasets remain, exacerbated by uncertainty among the research community as to what constitutes best practices. As an interdisciplinary team with diverse data management experience, we recognize the growing need for guidance on comprehensive data management practices that minimize the risks of data loss, maximize efficiency for stand-alone projects, enhance opportunities for data reuse, facilitate Indigenous data sovereignty and uphold the FAIR and CARE Guiding Principles. Here, we describe four fictional personas reflecting differing user experiences with data management to identify data management challenges across the biodiversity genomics research ecosystem. We then use these personas to demonstrate realistic considerations, compromises and actions for biodiversity genomic data management. We also launch the Biodiversity Genomics Data Management Hub (https://genomicsaotearoa.github.io/data-management-resources/), containing tips, tricks and resources to support biodiversity genomics researchers, especially those new to data management, in their journey towards best practice. The Hub also provides an opportunity for those biodiversity researchers whose expertise lies beyond genomics and are keen to advance their data management journey. We aim to support the biodiversity genomics community in embedding data management throughout the research lifecycle to maximize research impact and outcomes.
1 INTRODUCTION
The field of biodiversity genomics has undergone a fast-paced transformation over the last decade. Once largely inaccessible for non-model organisms, advancements in sequencing technology have substantially reduced costs associated with generating these data, leading to significant increases in the types and volumes of genomic data. Today, biodiversity genomics is a highly dynamic research field that integrates methods pioneered in human health (e.g., genome-wide association studies; Ozaki et al., 2002), agricultural breeding programmes (e.g., inbreeding coefficients; Wright, 1922) and principles from molecular ecology and evolution (e.g., identifying the genomic consequences of small population size; Duntsch et al., 2021; Khan et al., 2021; Liu et al., 2021; Robledo-Ruiz et al., 2022). The proliferation of this Digital Sequence Information (DSI) and related data is being utilized to address an ever-expanding array of research questions with wide-ranging potential benefits across society and is a challenge for existing data management systems and research community practices.
To maximize the short- and long-term impacts of biodiversity genomic data, a considered and careful approach to data management is essential. Good data management practices (Box 1) can benefit research teams and institutes, the research community and wider society when biodiversity genomics data is used to address contemporary socio-environmental challenges. For research teams, the positive impacts of data management can be particularly pronounced for large and long-term projects where there is regular turnover of members and/or research roles are highly partitioned. Effective data management benefits research teams through ensuring efficient resource use (e.g., time, computational and financial), risk mitigation (e.g., data loss, misinterpretation and misuse), signalling credibility through data reproducibility (Baker, 2016; Eisner, 2018) and ease of data-sharing for enhanced collaboration (Lau et al., 2017; Möller et al., 2017; Riginos et al., 2020). For research institutes and/or funding organizations there may be legal obligations and long-term responsibilities (including social licence requirements) for them as custodians to maintain the integrity of research data. Furthermore, these information-rich biodiversity datasets have immense reuse value that can only be realized if the data-generating researchers/institutes undertake careful data management (Crandall et al., 2023; Toczydlowski et al., 2021). These secondary use cases may diverge from the original purpose of data generation (Hoban et al., 2022; Leigh et al., 2021) and can provide additional valuable insights (e.g., Crandall et al., 2019), enhancing the value of these data to the research community and their potential impacts on society (e.g., Beninde et al., 2022; Exposito-Alonso et al., 2022).
BOX 1. Best practices versus good practices.
Based on our lived experiences working in this field, we (the authors) recognize there are different standards of data management. We acknowledge that achieving best practices (i.e., those described in the community guidelines and standards we strive towards implementing) is aspirational and may not always be practicable within the constraints of a research project (see section Exploring biodiversity genomic data management challenges). Instead, we encourage researchers to pursue “good practices” as a stepping-stone on the journey towards best practices.
In our own data management journeys, we have experienced situations where there has been little to no data management throughout the research lifecycle. For example, when tracking and troubleshooting code as new PhD students, as postdoctoral researchers attempting to standardize data storage and handling practices within research groups and as research team leaders working to ensure continuity within and across projects. Through our collective hindsight one lesson is clear—that any data management is better than no data management.
A lot of trouble can be saved by reaching out for advice and guidance about specific needs (even when unsure of what these are) from eResearch support staff early and often. We strongly encourage any incremental improvements to data management by individuals, as capacity allows. This may include gradual updates to established protocols, rather than attempting a hasty overhaul that you, or your colleagues, may not have the capacity to execute well. We also recognise that the culture of biodiversity genomics research is changing, and data management practices today may not mirror those of the past. Rather than lamenting past inadequacies, we encourage future-focussed data management solutions. These may include incrementally building data management habits into daily work and starting conversations among team members about their data and how they keep track of it. Together, these actions can go a long way towards shifting mindsets and propelling people along their data management journeys.
The incentives to implement data management practices are clear, and although there exists conceptual guidance on best practices within the broader scientific community (e.g., the FAIR Guiding Principles for scientific data management and stewardship, Wilkinson et al., 2016; and the CARE Principles for Indigenous data governance, Carroll et al., 2020, 2021; Jennings et al., 2023), implementation remains challenging (Box 2). Contributing factors include the sheer volume of these information-rich datasets and the associated resource requirements (i.e., the time and financial costs of data curation, maintenance and processing; Batley & Edwards, 2009; Chiang et al., 2011; Grigoriev et al., 2012; Schadt et al., 2010), as well as the inability of existing data standards, infrastructures and repositories to keep pace with the changing needs of this research community (e.g., Crandall et al., 2023; Liggins et al., 2021). Best practices for biodiversity genomic data management are an active area of discussion among the biodiversity genomics community (Anderson & Hudson, 2020; Fadlelmola et al., 2021; Field et al., 2008; Liggins et al., 2021; Yilmaz et al., 2011). However, these initiatives can be easily missed by biodiversity genomics researchers because they are often disseminated as discipline-specific outputs (e.g., publications, conference presentations and blogs) or institute-specific internal documents. This is further compounded by the absence of broad community standards administered by funding bodies and institutes. Thus, there are opportunities to centralize these existing resources. There are also benefits for research teams in extending their networks beyond the biodiversity genomics community to leverage the wealth of knowledge available across disciplines and institutes (e.g., information technologies [IT], data science and human genomics).
BOX 2. Ethical considerations for biodiversity genomic data management.
The potential for data misuse (e.g., cherry-picking, data theft, unpermitted use, sharing, or misappropriation) is ever-present throughout the data lifecycle (Cragin et al., 2010). Data misuse is harmful to the integrity of the research, science and innovation sector, and has important social implications due in part to an erosion of public trust in science (Laurie et al., 2014). Misuse can have direct negative impacts for participants, communities, research partners and end-users who may miss out on benefit-sharing as a consequence (a goal described in the Kunming-Montreal Global Biodiversity Framework, including for DSI; https://www.cbd.int/decisions/cop/?m=cop-15). This harm can further extend to the research team, collaborators and their institutes in the form of serious legal implications, reputational risk and negative impacts on career trajectories. There are clear ethical processes for other aspects of research (such as regulatory bodies for human and animal ethics) but such ethical frameworks may not yet be established for the generation and storage of biodiversity genomic data (especially for rapidly developing tools such as environmental DNA).
Data management is a tool researchers can use to mitigate these risks and some institutes and communities are well-versed in defining and implementing consistent and effective data management practices. However, we recognize that there remain gaps between knowing and doing, with different groups positioned at different points on their data management journeys. Nonetheless, good data management minimizes the risks of data misuse, loss, or theft, improves transparency and ensures data FAIRness within established parameters specific to those data.
Data management practices also seek to find a balance between “Open Data” and “Accessible Data”, the latter of which may be more appropriate for data pertaining to species and locations significant to Indigenous Peoples (e.g., Henson et al., 2021; Rayne et al., 2022; Te Aika et al., 2023). To facilitate Indigenous data sovereignty, open data should be accompanied by metadata that includes details of appropriate permissions, which may include access restrictions. Local contexts notices and biocultural labels offer one such framework to support this (Anderson & Hudson, 2020; Liggins et al., 2021).
By necessity, biodiversity genomics brings together diverse teams with broad interests. In this perspective, we aim to support biodiversity researchers, especially those with genomics expertise (i.e., data management practitioners), in embedding data management throughout the research lifecycle. We are a cross-institutional, interdisciplinary, multi-career stage collaborative team based in Aotearoa New Zealand, including biodiversity genomics researchers (N.J.F., J.W., L.L., T.E.S.), institutional and national eResearch and libraries staff (A.A., F.B., J.H., D.S.) and researchers with experience in being responsive to Indigenous considerations pertaining to culturally significant biodiversity genomic data, both as Indigenous (M.H.) and non-Indigenous scholars (N.J.F., J.W., L.L., T.E.S.). We have lived experience with the caveats of applying data management theory to real-life research situations, through starting from scratch with new projects and minimal prior experience of data management, inheriting existing data sets that require careful curation and adapting to a rapidly developing field where data types and associated data management practices have altered dramatically. Our extensive experience includes overseeing biodiversity genomic research projects, curating and managing biodiversity genomic datasets, developing project-specific data management plans (DMPs) and providing data management solutions to research teams, and much of this includes working with culturally significant data sets (e.g., Forsdick et al., 2021; Liggins et al., 2021; Magid et al., 2022; Rayne et al., 2022; Te Aika et al., 2023; Wold et al., 2023).
- describing typical data management experiences of individuals across the research ecosystem.
- presenting solutions to the questions and challenges that may arise when documenting and managing genomic datasets, and suggesting simple tools to support researchers in adhering to the FAIR and CARE Guiding Principles.
- creating the Biodiversity Genomics Data Management Hub (http://genomicsaotearoa.github.io/data-management-resources/) which contains curated resources including guidelines and standards for data management, along with tips and tricks that can be readily adopted and/or adapted for wide usage in biodiversity genomics projects.
We encourage researchers to view data management practices as behaviours intrinsic to the research process, and to adopt a mindset of adaptability to the various hurdles that may be encountered along the way. Through sharing these perspectives, we hope to support emerging researchers and the biodiversity genomics community more broadly on their data management journeys, and ultimately to amplify the real-world impacts of biodiversity genomics research.
2 EXPLORING BIODIVERSITY GENOMIC DATA MANAGEMENT CHALLENGES
Here, we present four fictional user experience personas to describe data management needs for individuals in different career stages and roles. These include a PhD student starting their project, a postdoctoral researcher working on long-established projects, a principal investigator seeking to facilitate research and an eResearch support staff member striving to support researchers. Using these personas, we aim to highlight some of the many important considerations associated with genomic data management. While we acknowledge that real life is not typically this tidy, we hope that researchers may see their own experiences reflected through some combination of these personas. The layers of challenges experienced by researchers may include the growing volume and types of genomic data and metadata, rapid technological and methodological advances, ensuring interoperability with metadata and balancing data openness and Indigenous data sovereignty needs.
2.1 Persona 1. A student new to biodiversity genomics
New PhD student Taylor Smith (Figure 1) has started a research project that will generate genomic data to inform conservation management for a culturally significant species (a recently described species of endemic lizard). Their project involves data collection and generation, analysis using the local compute infrastructure provided by their institute and dissemination of results to end-users including conservation practitioners and local communities. They will be operating under a DMP adapted from the template used across their research team, and they have access to internal training and external support structures.
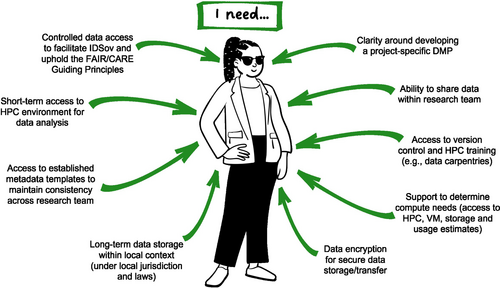
Their research team is in the process of developing a research manual that includes daily data management processes, along with on/offboarding procedures. Taylor is grateful for the supportive research environment, as they feel comfortable asking questions and sharing thoughts to help develop these processes. They are aware through conversations within their PhD cohort that this is not the situation for everyone. While their data is yet to be generated, being involved in these processes ensures they have a clear understanding of what will be involved in managing their data.
The primary challenges Taylor faces are in ensuring their data management practices facilitate Indigenous data sovereignty and uphold both the FAIR and CARE Guiding Principles during the active life-span of the project. To achieve these aims, they are relying on the guidance of existing frameworks (e.g., Collier-Robinson et al., 2019; King & Steeves, 2023; McCartney et al., 2023), and are well-supported in this by their research team leader, Professor Nepia (Persona 3) and the wider team. As the project has a defined end date, they also want to ensure that there is a framework in place to maintain these practices into the future. Communication around data management is primarily with Professor Nepia, who maintains trust-based relationships with the Indigenous Peoples that have strong cultural ties to the focal species, with support from eResearch and libraries staff at their institute.
2.2 Persona 2. An early career researcher working collaboratively outside of academia
Dr. Atsushi Sato (Figure 2) is a postdoctoral researcher at a national research institute, and contributes to several large international biodiversity genomics collaborations (including with Professor Nepia, Persona 3). These projects vary in scale, longevity and data management requirements. Each project Dr. Sato is involved with has its own established DMP, so he must take care to ensure that the workflows he uses for each project align with the respective DMP. Although he has some input in research planning and dissemination of results, his primary focus is on the analysis of large datasets, and specifically in incorporating environmental and climate data alongside genomic data. To do this, he relies on comprehensive and consistent metadata for each dataset.
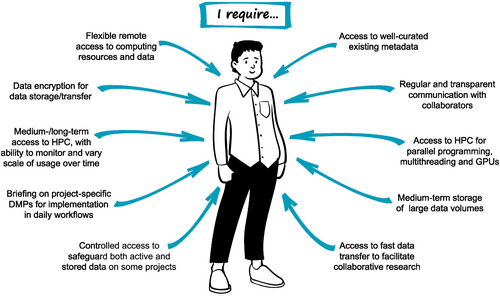
He is experienced in biodiversity genomics, and is able to clearly report his data management needs to eResearch and libraries staff at his research institute. These needs predominantly relate to short-/mid-term storage and access, as the long-term storage of most of the datasets Dr. Sato works with is the responsibility of researchers at other institutes. Dr. Sato also receives support from eResearch staff that deliver the national high-performance computing (HPC) infrastructure, where he can harness multithreading and parallel processing for analysing these large datasets.
Among the collaborators Dr Sato works alongside, there is a range of data literacy and data management experience, which can create communication challenges. He is aware that some data he has inherited was generated prior to the development of practices including Indigenous consultation and engagement and data sovereignty for culturally significant data. His knowledge of the shift in perspectives around these factors results in friction when he has made suggestions regarding the inclusion of these aspects in DMPs, and he is aware that publication of these data may be challenging due to the changes in journal publishing requirements. However, he views these issues as the responsibility of the collaborator who has led this project since its inception.
While Dr. Sato's skills are in high demand, he has been persistently employed on precarious short-term contracts. He finds this stressful, and is constantly looking for new opportunities that may propel him towards his goal of attaining a permanent research position. These concerns impact his research priorities, as he perceives trade-offs between time spent on data management and that spent on data analysis that can produce results that contribute towards his publication record. He is unwilling to risk conflict with his collaborators over the inclusion of data sovereignty and Indigenous engagement, as he fears that conflict may jeopardize his career prospects. From Dr. Sato's perspective, data management is an onerous task.
2.3 Persona 3. A biodiversity genomics research team leader
Professor Tehara Nepia (Figure 3) is a principal investigator at a university overseeing a conservation genomics research team including postgraduate students (including Taylor Smith, Persona 1), postdoctoral researchers and research associates (including Dr Atsushi Sato, Persona 2). Her focus is on designing, facilitating and disseminating research, and providing a supportive environment that produces highly skilled emerging researchers well-equipped to contribute to the research, science and innovation sector. Professor Nepia also places a strong emphasis on building and maintaining trusted relationships with research partners, including Indigenous Peoples. A substantial part of her role includes seeking and managing resources (including funding, computational resources and data storage) for the research team.
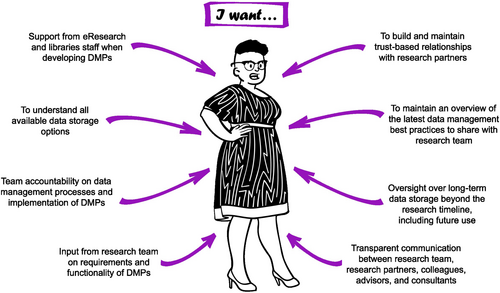
As the volume of data generated by Professor Nepia's team is continually expanding, there is a growing need to ensure a smooth transition of data (including metadata) between members of her research team. She has observed extensive change in data types and their associated data management practices during the course of her career. Professor Nepia has a responsibility to meet institutional requirements, and she is also committed to embedding data management practices that facilitate Indigenous data sovereignty and uphold the FAIR and CARE Guiding Principles.
Professor Nepia is working towards establishing a DMP template for use across all her research team's projects. To achieve this, she encourages open two-way communication with her research team to gain their perspectives of the needs and challenges associated with data management. She relies upon her research team to adhere to the DMPs, to support and encourage each other to do this, and to seek strategic advice from her when needed. Beyond the DMPs, Professor Nepia and her team co-develop research group guidelines that include data management practices to streamline team on/offboarding, allowing new members to quickly get up to speed and providing clear expectations of data management for those departing. Challenges may arise if she finds research team members becoming disengaged or unwilling to prioritize data management, so she needs to be able to pick up on these signals quickly and provide the necessary support.
Professor Nepia also engages with colleagues in similar situations nationally and internationally, including her disciplinary research community. Keeping abreast of evolving best practices in the biodiversity genomics research community and updating the research team's DMP template accordingly is an added pressure on her limited time; she never feels completely up-to-date with the latest developments but understands she must be the one in the research team to lead data management practices even if she is only able to support “good” versus “best” practice (Box 1). To help with this burden, Professor Nepia prioritizes building strong relationships with local eResearch and libraries staff (including Darryl Baker, Persona 4) that are based on transparent, timely, bidirectional communication. Through knowledge-sharing, eResearch and libraries staff help her to understand local data management capacity and constraints, and gain the necessary understanding of the project-specific nuances that enable the delivery of wrap-around solutions that support the needs of the research team now and into the future.
2.4 Persona 4. An eResearch staff member
Darryl Baker (Figure 4) is an eResearch Manager at a university, and provides eResearch support to numerous research projects across all disciplines and departments, including providing advice and services relating to compute and data storage facilities for biodiversity genomic data. Darryl recognizes how fortunate he is to be employed at an institute that recognizes the value of eResearch staff and the need for consistent data management practices, and that his team is sufficiently resourced to provide the support required by researchers. Darryl manages the resource that is the institutional compute and storage facilities allocated to research. He keeps up to date with research-focused technologies, consults with research teams and mentors researchers on the use of the available research systems. Over the past 4 years, the storage facility of the institution has reached peak capacity, requiring careful resource management. Darryl seeks budget approval to expand the current on-premise storage facility. Based on quotes provided by vendors, purchasing additional storage infrastructure proves to be expensive. Further, it will only provide a short-term fix as the institution's research data is predicted to exceed the storage limit within 5 years.
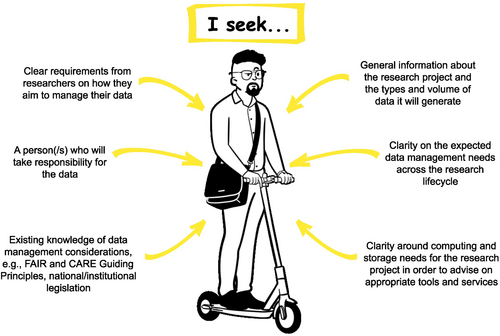
Recently, Professor Nepia (Persona 3) reached out to Darryl for eResearch services and support for her biodiversity genomics research team. Professor Nepia's team generates a number of projects, with rapidly increasing data management needs over the last 10 years. Darryl meets with one of Professor Nepia's research students, Taylor Smith (Persona 1), to understand the eResearch needs of an upcoming project about a new species of lizard. During the meeting, Darryl gathers information about the data being produced. Early indications are that this project will generate vast amounts of data and function under a DMP. Darryl wishes to understand the project-specific needs in order to advise on appropriate storage and computing solutions that will facilitate Indigenous data sovereignty and uphold the FAIR and CARE Guiding Principles. Darryl holds a clear understanding of the constraints arising from the institutional infrastructure and the responsibilities of the researcher under national and institutional legislation. Through conversations with researchers and research teams, Darryl can gain a clear vision of what they are trying to achieve within these constraints and provide advice and solutions to overcome data management pain points that may arise.
3 ADDRESSING THE CHALLENGES
Following the description of these personas, it is clear that while each persona will experience unique challenges, they also share common ones such as institutional support (e.g., the provisioning of institutional guidelines and policies pertaining to data management) and resourcing (e.g., time, funding allocations and access to data storage solutions). Here, we acknowledge the typical lag period between users identifying their own needs, institutional recognition of the broad nature of these needs and the subsequent provisioning of resources (e.g., the development of guidelines/policies, infrastructure and funding) to support these needs.
3.1 Resources to support researchers in implementing effective data management
To reduce the frustration often experienced by researchers on their journey towards best practices in data management, we have created the Biodiversity Genomics Data Management Hub (https://genomicsaotearoa.github.io/data-management-resources/). In the Hub, we identify key questions from the personas during their data management journeys based on existing challenges and uncertainties within the system, and connect these to modules that provide topic-specific tips, tricks and resources, including from beyond the traditional biodiversity genomics literature (Figure 5).
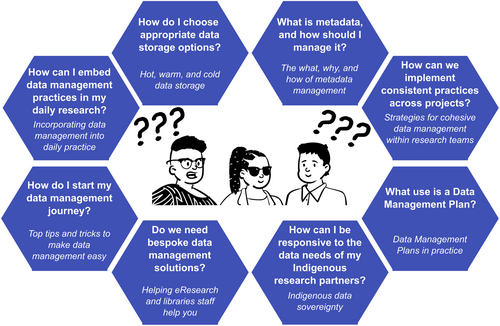
Module content draws on the diversity of our experiences and knowledge, with topics including: “Hot, warm, and cold data storage”, “Data Management Plans in practice”, and “Helping eResearch staff help you”. These tips and tricks are largely hard-won through the trials and tribulations experienced during our personal research journeys. We intend for the Hub to be a living resource that evolves over time, incorporating new tools and practices as these come to light. We welcome suggestions of additional module topics, along with contributions of the latest resources via the associated GitHub “Issues” page for feedback and discussion. We envision that the Hub will be of special interest for emerging researchers, and will be useful as a teaching resource, instilling data management practices as part of daily workflows from the beginning of the research journey. The Hub may also provide an opportunity for those with an interest in data management outside of the biodiversity genomics space to have the opportunity to peek “through the looking glass” and gain insight into the similarities and differences with their own fields. In assembling resources for the Hub to address challenges across personas, three overarching actions stood out as immediately accessible steps towards best practices for the biodiversity genomics community. Here, we elaborate on these.
3.2 Develop data management plans
Biodiversity genomic data management tends to come into focus at the end rather than throughout the research lifecycle. Many journals that publish biodiversity genomic research have open data policies (e.g., the Joint Data Archiving Policy), and this may be the first instance at which researchers are required to demonstrate data management. Indeed, genomics broadly appears immature compared with other disciplines in terms of data management (e.g., data science, IT and human genomics). For example, DMPs are often perceived as “nice to have” but are not yet widely required. However, when working with the large volumes of data produced via genomic sequencing, and/or in research teams distributed across multiple institutes, data management can quickly degenerate leaving the data, researchers and research partners vulnerable (Box 2). Further, DMPs are one tool among many that will be required to achieve the benefit-sharing goals pertaining to genomic data as described in the Kunming-Montreal Global Biodiversity Framework (Decision 15/4: recognizing the contributions and rights of Indigenous communities and Decision 15/9: the generation, access and use of digital sequence information; https://www.cbd.int/decisions/cop/?m=cop-15).
DMPs are key tools for mitigating the risks of data loss and misuse. Where they do not already exist, we anticipate a widespread shift towards the establishment of data management policies within institutes and by research funding organizations (including the requirement of DMPs in research funding applications) in the near future (Bloemers & Montesanti, 2020; Fadlelmola et al., 2021; Jorgenson et al., 2021). Indeed, the primary research funding body in Aotearoa New Zealand, the Ministry of Business, Innovation and Employment, is shifting towards an open research policy (https://www.mbie.govt.nz/science-and-technology/science-and-innovation/agencies-policies-and-budget-initiatives/open-research-policy/) as many of its contemporaries have done (e.g., the Australian Research Council, the European Research Council, the National Institutes of Health), which may come to include a requirement for DMPs. We foresee that some of the challenges associated with requirements to provide DMPs during funding applications will be in ensuring cohesive frameworks for the development of DMPs that are fit for purpose and more broadly in the development and maintenance of trustworthy digital repositories at scale (Lin et al., 2020).
The inclusion of an approval and/or compliance pathway may be recommended to ensure that DMPs lead to meaningful actions in the improvement of data management in biodiversity genomics rather than simple “box-ticking” or thought exercises. Specifically, approval pathways could require consideration of the DMP during the funding application process to determine whether it is fit for purpose. In comparison, a compliance pathway could require researchers to demonstrate that data management actions have been carried out in accordance with the DMP provided. DMP approval and compliance regarding the FAIR Guiding Principles would require consideration by external assessment panels with discipline-specific knowledge and expertise. For data and metadata associated with species or locations significant to Indigenous Peoples (Box 2), decisions around auditing and assessment of DMPs in relation to the CARE Guiding Principles can only be made by the associated Indigenous Peoples. Indigenous leadership across the research ecosystem, including professional and research staff, will be essential in the co-development of any such systems, with one important consideration being ensuring that DMPs are responsive to current contexts while remaining flexible for the future. Indeed, there will not be a “one size fits all” solution for culturally significant data. We note here that supporting Indigenous research partners through the provision of adequate resourcing to engage with these processes will be essential (Te Aika et al., 2023).
While compliance is one method of ensuring that data management actions are implemented, research projects tend to change course over time, and a DMP designed during the planning stage may not provide the flexibility required to meet changing data needs later in the research lifecycle. Rather than using approvals or compliance processes to ensure appropriate data management actions are taken, a more feasible approach could be to recognize a DMP as a live document throughout the research process, allowing for updates as the project changes. In this scenario, version control methods should be used to track changes throughout the project. During any process of revision of the DMP, it will be important to maintain regular and transparent communication with research partners, to ensure that proposed changes are fit for purpose, while continuing to accommodate the needs and interests of all parties. At the end of the project, the research team could complete a self-reflective retrospective process, identifying which aspects went according to plan, where needs changed over time, and whether there were any limitations or challenges due to institutional or infrastructure constraints. This could help researchers to better understand the capabilities and capacities of their teams and systems, and inform future research design that includes DMP development. Further, sharing the outcomes of such retrospectives with associated eResearch and libraries staff will help to close the loop.
3.3 Seek support from eResearch and libraries staff
We challenge researchers to look beyond their immediate research community for assistance—help may be closer at hand than expected. Here we highlight the benefits of engaging with eResearch and libraries staff within or beyond your institute from an early stage in the research lifecycle. These professional staff are a supporting network holding knowledge and expertise in crafting solutions to data management challenges (Andrikopoulou et al., 2022). Researchers benefit from developing these relationships with staff who cultivate institutional knowledge and solutions that may not be captured in the traditional or domain-specific scientific literature. eResearch and libraries staff can provide guidance and targeted support in the co-development of project-specific data management strategies that include institutional operating requirements and the capacity and capability of existing infrastructure, and in incorporating data management practices into day-to-day research workflows.
eResearch and libraries staff may at times be overlooked due to the frequent tangible and intangible siloing of disciplines, resulting in researchers being unaware of how these staff can provide support, and unclear as to what their mandates are, with eResearch and libraries staff consequently unaware of the data management needs and challenges experienced by research teams. Further, eResearch and libraries staff are often spread thinly within institutes, with high demand for their services but limited capacity to provide much-needed support. As such, building channels of communication between research teams and support staff is key, and both parties must be willing to come to the table to share and learn from one another.
Developing strong working relationships requires reciprocity, with an emphasis on mutual benefit (which may include academic acknowledgement) and respect for expertise on both sides. eResearch and libraries staff often require knowledge of the research context and learned experiences from researchers so they can provide and/or procure the necessary services and support, and researchers can also endeavour to engage with the technicalities and concepts necessary for full and fruitful discussions. We recommend that researchers meet early and often with eResearch and libraries staff to discuss their data management needs. Investing in these relationships ultimately means that researchers will get the wrap-around support they require, and eResearch and libraries staff will be kept appraised of the changing needs of researchers, facilitating the development of future-focussed solutions.
3.4 Establish a research data management culture in your team
It is vital to ensure the continuity of data management throughout the research lifecycle. We strongly encourage researchers to step up and take an active leadership role in situations where there is an absence of clear and consistent guidelines. However, data management is most effective when pursued as a team, with a consistent and cohesive plan and division of labour. A little effort early in the process can go a long way, and so we recommend that research teams work together to develop clear documentation around on/offboarding procedures and daily data management practices. This will streamline the process of joining the team, provide guidance on the options for and constraints around data transfer, storage and access, and a clear pathway to follow when departing that may include ongoing access to data or the packaging of data and metadata for long-term storage.
As the importance of data management becomes increasingly recognized, but prior to the establishment of institutional roles, we envision an opportunity to create a new role within research teams—that of data management champion. We perceive such a role to be analogous to that of a lab manager, providing support and oversight for research teams across all aspects of data management. This role can ensure consistency despite the potential for frequent turnover within research teams through overseeing the onboarding and training of new members and ensuring the implementation of consistent data management practices across the research team. While anyone can take on this transferable role, a data management champion will ideally have a mid- to long-term position within the research team, hold a deep understanding of the unique characteristics of each research project and have the necessary level of autonomy to operate independently as a leader in this role. The data management champion can also operate as a conduit between the research team and eResearch and libraries staff, and so excellent people skills will be advantageous. By engaging regularly and often with their institute's support structures, they can ensure that eResearch and libraries staff are kept up to date with the changing needs of the team and ensure access to the latest services and support.
Given the importance of such a role, succession planning will be essential to ensure consistency and continuity for the research team. While we are currently aware of few research teams that have a data management champion, we perceive this as a “next step” in the community's collective data management journey. We emphasize the need for such a role to well-resourced, to avoid burdening individuals with additional (unpaid) responsibilities that may detract from their personal research trajectories. Further, we consider that the responsibilities delivered in this position will be highly transferable and sought after. For some researchers, this may be a step towards taking up other management responsibilities or roles in the future.
4 CONTINUING THE DATA MANAGEMENT JOURNEY
Here, we have presented tips and tricks to support biodiversity genomics researchers in the development of good data management practices, though we emphasize that any data management is better than none. Data management is a journey, and we are all on an aspirational path striving towards best practices. We trust our contribution, both here and in the Biodiversity Genomics Data Management Hub, will be a helpful guide for researchers new to biodiversity genomics, and a useful prompt for existing researchers to start data management planning early in the research lifecycle (e.g., when writing proposals) and to embed good data management practices into their daily research routines. Further, we are confident this contribution demonstrates the need for data management infrastructure and practices to be included as key aspects of the research lifecycle that require designated resourcing and support from institutes and funding bodies across a broad range of disciplines.
Glossary
- Accessible data. Data accessible under well-defined conditions, as per the FAIR Guiding Principles (Mons et al., 2017; Wilkinson et al., 2016).
- CARE Principles for Indigenous Data Governance. Designed to complement the FAIR Guiding Principles, these people- and purpose-oriented principles and supporting concepts (collective benefit, authority to control, responsibility, ethics) reflect the crucial role of data in advancing innovation, governance and self-determination among Indigenous Peoples (Carroll et al., 2020, 2021). https://www.gida-global.org/care.
- Data lifecycle. The steps in the research process specifically pertaining to data, from planning, collection and generation, analysis and collaboration, evaluation, storage, dissemination, access and reuse, which can contribute to the planning for new data generation. The data and research lifecycles are distinct but interrelated.
- Data management. The processes and practices associated with the documentation and storage of and access to data and associated metadata throughout the research lifecycle.
- Data management plan (DMP). A document describing the data that will be generated during a research project, and how it will be used, accessed, and stored during the research lifecycle. Also known as a data management and sharing plan, though in our definition of data management, data sharing is inherently included in data access.
- eResearch. The use of digital tools and techniques to advance research.
- eResearch and libraries staff. A broad group that includes research software engineers, research infrastructure developers, data scientists, data stewards and other professional services staff that deliver library, IT, bioinformatics and high-performance computational support.
- FAIR Guiding Principles. Guidelines for scientific data management and stewardship intended to improve the findability, accessibility, interoperability and reuse of digital assets (Wilkinson et al., 2016). https://www.go-fair.org/fair-principles/
- Indigenous data. The tangible and/or intangible cultural materials, belongings, knowledge, digital data and information about Indigenous Peoples or that to which they relate (Lovett et al., 2019; Rainie et al., 2019).
- Indigenous data sovereignty. The expression of a legitimate right of Indigenous Peoples to control the access, the collection, ownership, application and governance of their own data, knowledge and/or information that derives from unique cultural histories, expressions, practices and contexts (https://localcontexts.org/indigenous-data-sovereignty/).
- Metadata. Data that provides information about other data. For biodiversity genomic data, metadata can provide information regarding context (e.g., taxonomic, spatial, temporal and associated permissions) as well as used technologies/methodologies.
- Open data. Data anyone can use and share, typically openly accessible and with an open licence.
- Research lifecycle. The steps in the process of scientific research from inception (research planning, design and funding) to completion (dissemination of results and real-world impact), which often leads back to development of new related projects. The research and data lifecycles are distinct but interrelated.
- Virtual machine (VM). A software-based computing system emulating that of a different physical machine, often used to run a different operating system than that of the primary system of the physical computer.
AUTHOR CONTRIBUTIONS
Natalie J. Forsdick, Jana Wold and Tammy E. Steeves conceived the research. All authors provided input into the research direction and contributed through robust discussion towards the development of the manuscript and the creation of the Biodiversity Genomic Data Management Hub. Jamie Hart provided illustrations. Natalie J. Forsdick and Jana Wold wrote the first draft of the manuscript and led the writing of subsequent drafts. All authors provided feedback and approved the final manuscript.
ACKNOWLEDGEMENTS
The authors wish to thank the following people for their thoughtful advice, insights and friendly feedback during the development of this project: Mik Black, Thomas Buckley, Eric D. Crandall, Manpreet Dhami, Tom Etherington, Leanne Elder, Stephanie Galla, Tipene Merritt and the University of Canterbury (UC) eResearch Co-Design Group, David Medyckyj-Scott, Nick Spencer, Matt Stott and the UC ConSERTeam. We acknowledge the support of Manaaki Whenua – Landcare Research (NF), Genomics Aotearoa (NF, LL, TES, JW), NeSI (DS), New Zealand's Ministry of Business Innovation and Employment (MBIE) Infrastructure Platform (TES, JW), the University of Canterbury (AA, FB, JH, TES, JW), Massey University (LL), and the University of Waikato (MH). Open access publishing facilitated by Landcare Research New Zealand, as part of the Wiley - Landcare Research New Zealand agreement via the Council of Australian University Librarians.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
BENEFIT-SHARING STATEMENT
Benefits generated: A cross-institutional, interdisciplinary research collaboration was developed with all collaborators included as co-authors. Benefits from this collaboration accrue through the provision of the Biodiversity Genomic Data Management Hub, which is shared as a publicly available web resource to support biodiversity genomics researchers in improving data management practices across the data lifecycle. This research is timely given predicted changes in research funding requirements to include data management plans.
Open Research
DATA AVAILABILITY STATEMENT
No data were produced or analysed in the development of this manuscript.




