Arthropods are kin: Operationalizing Indigenous data sovereignty to respectfully utilize genomic data from Indigenous lands
Abstract
Indigenous peoples have cultivated biodiverse agroecosystems since time immemorial. The rise of metagenomics and high-throughput sequencing technologies in biodiversity studies has rapidly expanded the scale of data collection from these lands. A respectful approach to the data life cycle grounded in the sovereignty of indigenous communities is imperative to not perpetuate harm. In this paper, we operationalize an indigenous data sovereignty (IDS) framework to outline realistic considerations for genomic data that span data collection, governance, and communication. As a case study for this framework, we use arthropod genomic data collected from diversified and simplified farm sites close to and far from natural habitats within a historic Kānaka ʻŌiwi (Indigenous Hawaiian) agroecosystem. Diversified sites had the highest Operational Taxonomic Unit (OTU) richness for native and introduced arthropods. There may be a significant spillover effect between forest and farm sites, as farm sites near a natural habitat had higher OTU richness than those farther away. We also provide evidence that management factors such as the number of Polynesian crops cultivated may drive arthropod community composition. Through this case study, we emphasize the context-dependent opportunities and challenges for operationalizing IDS by utilizing participatory research methods, expanding novel data management tools through the Local Contexts Hub, and developing and nurturing community partnerships—all while highlighting the potential of agroecosystems for arthropod conservation. Overall, the workflow and the example presented here can help researchers take tangible steps to achieve IDS, which often seems elusive with the expanding use of genomic data.
1 INTRODUCTION
Global ecosystems are under siege, with threats to biodiversity approaching critical tipping points. Key native taxa are disappearing (de Oliveira Roque et al., 2018), while introduced species are increasingly spreading and disrupting the functioning of ecosystems (Burnett et al., 2006). Recognition that landscapes have been anthropogenically shaped for tens of thousands of years has directed efforts to understand biodiversity patterns beyond ‘pristine, wild and natural’ environments. Indigenous-managed agroecosystems have been noted for their capacity to maintain and bolster biodiversity (Perfecto et al., 2019). For Indigenous peoples, the culturally relevant biodiversity fostered by agroecosystems has shaped their identity and culture (Nelson & Shilling, 2018). Biodiversity conservation efforts have been vital to global indigenous sovereignty movements that seek to rematriate and restore lands where biodiversity has been eroded due to colonization (Settee & Shukla, 2020; Wezel et al., 2009). However, the historical and ongoing extraction of biodiversity resources with little engagement or benefit to communities has undermined Indigenous sovereignty and caused a rightful distrust by Indigenous peoples concerning biodiversity conservation studies and initiatives (Merson, 2000).
The growing popularity and utilization of metabarcoding and environmental DNA (eDNA) in biodiversity research has expanded the scale of data generation from Indigenous lands (Arribas et al., 2022; Kennedy et al., 2020). Moreover, in the case of many eDNA samples, a full new understanding of ecosystems can be gained in covert ways: A single water sample from a river can determine if a prized riparian species is upstream (Rees et al., 2014) or a bag of tea leaves bought from a supermarket can illuminate arthropod community composition (Krehenwinkel et al., 2022). The novelty, scalability, and covertness of eDNA-based data stress the need to understand how to respectfully use genomic data collected on Indigenous lands to support communities better and honor their sovereignty.
Consequently, efforts are underway at multiple governance scales to empower communities and protect Indigenous data sovereignty (IDS). For example, the United Nations Declaration on the Rights of Indigenous Peoples (UNDRIP) (Assembly, 2007) and Convention on Biological Diversity and its Nagoya Protocol (Convention on Biological Diversity—Article 1. Objectives, n.d.) all affirm Indigenous peoples have bona fide sovereignty that must be honored. Data collected within Indigenous homelands should be under the authority of relevant communities or an entity designated by each community. Initiatives such as the global Indigenous data alliance (GIDA) (Global Indigenous Data Alliance, n.d.) promote the exercising of IDS through the CARE (Collective benefit, Authority to control, Responsibility and Ethics) principles (Carroll et al., 2022) for Indigenous data governance. These human-centric principles sit alongside the more data-centric FAIR principles (Wilkinson et al., 2016) and guide researchers in operationalizing IDS through collective benefit, authority to control, responsibility, and ethics.
Global biodiversity genomics initiatives are beginning to recognize the importance of Indigenous peoples in their mission to sequence all of eukaryotic life (Mc Cartney, Anderson, et al., 2022; Mc Cartney, et al., 2023). Indigenous communities are also taking agency over their data by providing guidelines for researchers. Indigenous peoples across the globe have developed codes of research conduct to gain and retain agency over their biodiversity resources, including Māori (Hudson et al., 2016; Stats, 2020), First Nation, Metis and Inuit Peoples (TCPS-2, 2014), the San community (Callaway, 2017), Aboriginal and Torres Strait Islander Peoples (Guidelines for Ethical Research in Australian Indigenous Studies, 2012) and Tribes across the United States (Carroll et al., 2022).
The breadth of emerging IDS initiatives highlights the importance and potential of halting extractive research practices in Indigenous communities. Nonetheless, to many communities and researchers, IDS still seems to be an elusive goal, especially in its application. In this paper, we build on a framework developed by Mc Cartney, et al. (2023) by applying it to an empirical case study of arthropod genomic data collected along a gradient of agricultural diversification and proximity to natural habitat on Hawai'i Island, Hawai'i (Figure 1a). The McCartney, et al. (2023) framework recommendations are guided by the CARE principles to work more justly with Indigenous peoples and local communities (IPLC). These recommendations fit into six steps. However, for our case study, we found it best to summarize and organize our IDS workflow into the three steps presented below (and illustrated in Figure 2).
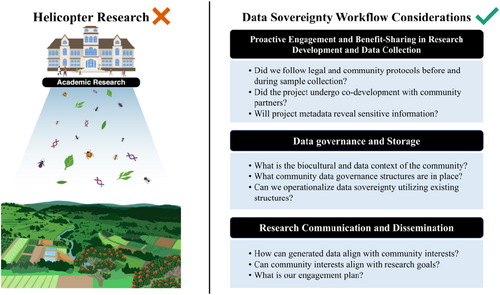
1.1 Proactive engagement and benefit-sharing in research development and data collection
Researchers must invest time and resources to develop relationships with community members to gain support and permission to access sites and obtain samples. Access should be obtained legally and ethically, such as those outlined by the Nagoya Protocol or following community protocols. Community partners should provide input and be a part of the co-development of project goals throughout the life of the study. It is critical to be transparent about initial project goals and benefits with community partners and the risks and benefits of storing samples away from the community's residence (if applicable). When curating metadata, researchers should redact specific sensitive metadata fields congruent with Dublin Core and Darwin Core (Wieczorek et al., 2012).
1.2 Data governance and storage
Researchers should understand and respect the Indigenous communities' cultural sensitivities, customs and protocols surrounding the governance of the data life cycle. A responsible data management plan should be developed to prioritize long-term sustained community access and perspectives.
1.3 Research communication and dissemination
Researchers should design a research communication and dissemination plan that considers a breadth of appropriate audiences, such as community members and partners, land managers, or researchers. Researchers should consider further project and partnership continuity through funding opportunities if possible and mutually desired.
1.3.1 Case study: ʻUpena of pilina: Revitalizing connections between Kānaka ʻŌiwi food systems and arthropods
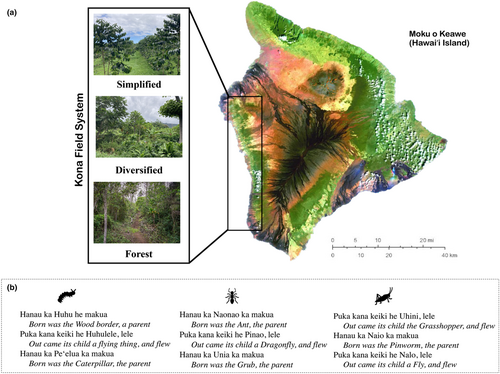
In Hawai'i, Kānaka ʻŌiwi (Indigenous Hawaiian; referred to as ʻōiwi hereafter) established vast agricultural systems spanning elevational ranges from the coast to upper-elevation mountainous areas (Kagawa & Vitousek, 2012; Lincoln & Vitousek, 2017). However, historical and ongoing colonization and globalization have drastically altered the agricultural landscapes of Hawai'i (Hutchins & Feldman, 2021). For example, in the Kona Field System (KFS), an agricultural belt built on the leeward side of Hawai'i Island, the proliferation of coffee caused the contraction of land dedicated to agroforestry practices and the introduction (both intentionally and unintentionally) of a myriad of invasive arthropods (Allen, 2001; Lincoln et al., 2018). Some introduced arthropods have even been linked to the decline of native and endemic arthropods (King et al., 2010) and flora (Roy et al., 2019), along with crop production (Messing, 2012).
More recently, a growing food sovereignty movement across Hawai'i has revitalized agroforestry practices and the subsequent return of ‘ōiwi community members and culturally significant species (Lincoln et al., 2018). In the KFS, after being abandoned for many years, agroforestry sites are being cultivated again by the ‘ōiwi community, where they are growing crops with known associations with native arthropod diversity (Swezey, 1954). Today, these traditional agroforestry sites are nested within a complex landscape mosaic dominated by conventional coffee monocultures. Yet, whether greater agricultural diversification can provide suitable habitat to sustain native biodiversity rather than serve as an avenue for the proliferation and spread of non-native species remains unresolved. Therefore, our ongoing research in the Kona Field System asks: to what extent can diversified agricultural landscapes support native arthropod diversity? How does arthropod community composition shift in response to crop diversification?
2 METHODS
Here, we present a simplified version of our methods. Please see Appendix S1 for more details on our methods.
2.1 Site selection and arthropod sampling
We selected six farm sites along a diversification gradient, which was based on the presence of on-farm crop diversity, size (area in production), elevation, and similar utilization of inputs such as pesticides and fertilizer. Due to our interest in comparing farm arthropod community composition and structure to that in forested areas, two forest sites were sampled. Forest sites were selected based on the degree of disturbance and elevation: primary forests facing degradation from invasive plants and arthropods located between 792 and 914 m in elevation. Therefore, forest sites had a mix of native and introduced vegetation. We also ensured that all sites were at least 800 m apart.
2.2 Sample collection
We collected arthropods using timed vegetation beating (40 seconds) at five points (2 m radius) along a 25-meter transect. Before beat sampling, we collected and sifted leaf litter from a 1 × 1 m plot at each transect point. Arthropods from litter samples were then collected using a Berlese funnel. All arthropod samples were stored in 95% ethanol at −20°C until we conducted DNA extractions.
2.3 DNA extraction
DNA extraction of size-sorted arthropod-plant community samples was performed using the Tissue protocol described in the Qiagen Puregene kit modified for automation (Lim et al., 2022).
2.4 Library preparation and sequence analysis
We used a primer combination (ArF1 - Fol-degen-rev) which targets a 418 bp fragment in the barcode region of the Cytochrome Oxidase I (COI) gene (Lim et al., 2022) in triplicate amplifications using the Qiagen Multiplex PCR kit (Qiagen). The three sample replicates were pooled, and the quality of all pooled PCR products was ensured through bead cleanup and fragment length analysis. Final amplicon libraries consisted of pooled equimolar samples and were sequenced on an Illumina® MiSeq.
Sequences were demultiplexed on Illumina® BaseSpace. PCR primers were trimmed using Cutadapt (Martin, 2011). Sequences were merged, filtered, and denoised to amplicon sequence variants (ASVs) using DADA2 (version 1.14.1.; Callahan et al., 2016; Brandt et al., 2021). ASVs were then clustered to 3% radius (97%) OTUs using DECIPHER (2.14.0; Wright et al., 2012). A curated OTU table was created using LULU (version 0.1.0; Frøslev et al., 2017; Brandt et al., 2021). All remaining OTU sequences were compared to Genbank using ElasticBLAST on Amazon Web Services.
2.5 Native/introduced assignment
To assign a native or introduced status to all OTUs, we utilized NIClassify (https://github.com/tokebe/niclassify), a software tool that implements a machine-learning strategy based on the principles of Andersen et al. (2019). The tool has been used to accurately assign status to several arthropod datasets from Hawai'i (Graham et al., 2022; Kennedy et al., 2022).
2.6 Agricultural diversification index
To create the agricultural diversification index, we used the first principal component of a PCA matrix that included the scaled values of all management attribute variables (coffee cover, crop diversity, non-crop vegetation, canopy layers, litter depth, and distance to natural habitat) (Lu et al., 2022; Armengot et al., 2011). The index allowed us to explore the overall effect of agricultural diversification.
2.7 Statistical analysis
We assessed the alpha-diversity (observed ‘richness’) and beta-diversity (‘composition’ based on Bray–Curtis dissimilarities of Hellinger-transformed community matrices) of native and introduced arthropods in two ways. First, we examined site-level differences between richness and composition to address farmer's interests, including the individual environmental and management attributes that drive these differences and the individual arthropod taxa that contribute to site-level differences. Next, we tested the effect of agricultural diversification (combining all management attributes into a singular index) and distance to natural habitat (‘close’ and ‘far’) to examine the ecological mechanisms that drive the richness and composition of arthropods.
2.8 Site-level differences in arthropod communities
2.8.1 Alpha-diversity
We examined the differences in observed native and introduced arthropod richness (Poisson error) between sites using generalized linear mixed models with site as a random effect using the lme4 and lmertest packages in R (version 4.2.3) (Bates et al., 2015; Kuznetsova et al., 2017; R Core Team, 2020). Tukey HSD's was also performed to observe pairwise comparisons between sites.
2.8.2 Beta-diversity
To determine the environmental and management attributes that significantly influenced introduced and native arthropod community composition, we used a distance-based redundancy analysis (dbRDA) using the vegan package (Oksanen et al., 2013). The dbRDA tests how much variation within a community (i.e. arthropod community composition) is explained by a group of explanatory variables (i.e. environmental and management variables) (Legendre & Anderson, 1999). Collinear variables were removed, and the significance of the coefficients was determined using a permutation-based ANOVA.
To understand the contribution of individual taxa to the dissimilarity between sites, we performed a similarity percentage (SIMPER) analysis on the introduced and native arthropod composition matrices.
2.9 Agricultural diversification and landscape effects on arthropod communities
2.9.1 Alpha-diversity
We tested the effect of agricultural diversification, distance to natural habitat (‘close’ vs. ‘far’), and their interaction on observed richness of introduced and native arthropods using generalized linear mixed models with site as a random effect using the lme4 and lmertest packages in R (version 4.2.3) (Bates et al., 2015; Kuznetsova et al., 2017; R Core Team, 2020).
2.9.2 Beta-diversity
To evaluate the effects of agricultural diversification, distance to natural habitat (‘close’ vs. ‘far’), and their interaction, on introduced and native arthropod community composition, we used a permutational multivariate analysis of variance (PERMANOVA) using the package vegan (Oksanen et al., 2013). PERMANOVA tests compositional differences by examining whether the centroids of sample clusters differ. To illustrate arthropod community composition differences for the interaction between agricultural diversification and proximity to natural habitat, the composition matrices were ordinated by a principal coordinates analysis (PCoA) using the ‘pcoa’ command in the ape package (Figure 1a) (Paradis et al., 2004).
2.10 Proactive engagement in research development and data collection
2.10.1 Positionality and motivation of researchers during engagement
Potential farmer participants were engaged through the University of Hawai'i Cooperative Extension, pre-established relationships, and farm visits. Through discussions with the farmers, the project team gained a vital understanding of the history of the land, farmer interests, and pest issues. Importantly, these discussions also allowed farmers to ask questions about the project and its design. Although it is a best practice to co-develop the project goals with the community, the study design occurred before engagement. However, through initial engagements and conversations, the project team acknowledged their position as researchers and recognized the power, potential harm, and responsibility of conducting research on these lands. These relationships made engaging from that point onwards and guiding future research plans possible. For instance, during a discussion with one farmer, they expressed a passion for aligning academic research with on-farm applications. This led to a grant application for a co-developed project with a community partner, Kamehameha Schools, along with farmer input from within the KFS, which was successfully funded. Notably, this farmer is a paid consultant on the project with several others. This demonstrates that engagement at any point in the project lifecycle is highly beneficial.
The positionality of the research team during these engagements with the community and landscape is essential to identify and honor to understand the power imbalances and differing perspectives that occur. Regarding our research team, the lead author is part of the ʻōiwi community. A certain unmeasurable level of interpersonal communication comes with holding this identity, including how to approach and interact with community members and social normalities within local communities in Hawai'i. However, the lead author and the research team acknowledged their position as researchers from an institution such as UC Berkeley that creates a power imbalance (please see Baum et al. 2006 for further scholarship on the role of identity in participatory research).
The motivation behind the inception and planning of a research project with community partners is important to recognize and acknowledge. Motivation brought on by romanticism, white savior complex, or a need to fulfill a grant requirement for broader impacts or a DEIJ (Diversity, Equity, Inclusion, and Justice) component can be common and do not create sustained and trustworthy relationships with communities. An example of romanticism is the desire to work in a location such as Hawai'i because of its beauty or historical public perception as a ‘paradise’ or to engage with an Indigenous community based on notions of needing to ‘save’ them from poverty or injustice. The motivation for our research comes from the lead author's long-term, sustained interactions with both landscape and people over many years. This built relationship and the positionality outlined in the section above created a kuleana (responsibility) to continue building and bettering these project relationships.
2.10.2 Safeguarding metadata and ex-situ samples
While sampling arthropods at each site, we scored or measured various environmental and management attributes (Table 1). These attributes were selected based on their known ability to shape arthropod communities. Metadata collected through the project discloses in-depth information about each farm site and the arthropods on them. All metadata identified as culturally sensitive by the community, such as location and identity beyond the order level of species, was redacted from publicly available metadata according to Darwin Core standards (InformationWithheld; Table 2). This ensures that the most sensitive data revealing the location of arthropod and plant species, along with their identity, will not be available to those outside of the community, thus reducing the ability for unauthorized visits or access. Farmers and community members involved in the project will have access to the complete, unredacted version of the metadata records about their site. Unredacted data sharing among farmers is facilitated through a case-by-case approval basis.
| Metric | Description |
|---|---|
| Coffee cover | The percentage of coffee cover at a site: no coffee (0), 25% cover (0.33), 75% cover (0.66), 100% cover (1) |
| Crop diversity | A score based on the number of different crops grown on a site: 0 crops (0), 1–2 crops (0.33), 3–8 crops (0.66), and 8+ crops (1) |
| Non-crop vegetation | A score based on the presence and taxonomic origin of non-crop vegetation: no non-crop vegetation (0), only non-native (0.33), both non-native and native (0.66), and only native (1) |
| Canopy layers | The sum of the presence (1) or absence (0) of different canopy layers on a site: herbaceous, shrub understory, lower canopy, upper canopy, and emergent |
| Litter depth | The average measurement of leaf litter depth at each sample collection point (centimeters) |
| Distance to natural habitat | The distance to the edge of the closest forest habitat measured on ArcGIS (meters). The range in distance for farm sites varied from 390 to 2188 m |
- Note: These attributes were selected based on their known ability to shape arthropod communities.
| Sample ID | Order | Genus and species | Location | rightsURL | rightsIdentifer |
|---|---|---|---|---|---|
| 1 | Diptera | Informationwitheld | Informationwithheld | https://localcontextshub.org/researchers/projects/33 | BC-Notice |
- Note: Many metadata standards, including the iBOL manifest we utilized, do not have fields to recognize the rights of Indigenous peoples. We also redacted sensitive fields according to Darwin Core standards.
Once collected, the samples were transported to UC Berkeley for processing, where DNA extraction, library preparation, sequencing and bioinformatics occurred. The gDNA from this project will be stored in a freezer on campus and not used for non-project purposes. The research team recognizes that the samples were processed far from their origin. All farmers were aware of the destination of the samples. In the future, there is tremendous potential for ʻōiwi geneticists and computer scientists to gather to create a laboratory and biobanking operation that is accountable to community standards, which could be modeled after the Native BioData Consortium, an Indigenous-led organization further discussed below (see Section 2.11.4).
2.11 Data governance and storage
2.11.1 Contextualizing community data
Due to colonial practices and policies, the research enterprise has resulted in most Indigenous data being generated and analyzed away from the origin. Systemic inequities and power imbalances perpetuate unjust disconnections between Indigenous communities, their samples, and data. In Hawai'i, previous and ongoing biopiracy projects, plant patenting, and human genome projects have caused a rightful hesitancy among the ‘ōiwi community concerning the genomic research enterprise (Goodyear-Kaʻōpua et al., 2014). In 2003, in response to the increasing commercialization, commodification, and exploitation of Indigenous resources, such as kalo (taro), ʻōiwi elders and cultural leaders crafted the Paoakalani Declaration (2003). This foundational document outlined ʻōiwi perceptions of traditional knowledge, genetic and biological material stewardship, and a governance framework (Figure 3).
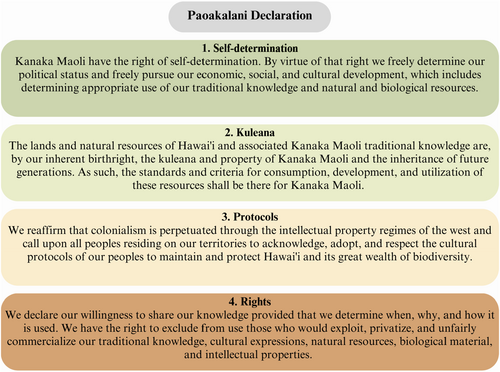
In this project, the arthropods we collected are biological and genetic material from Hawai'i. Therefore, they are protected under the Paoakalani Doctrine. Moreover, culturally, arthropods are kin to ʻōiwi. Several species—both native and non-native—are mentioned in the Kumulipo (Figure 1b), the Hawaiian life origin story. The presence of arthropods in the Kumulipo ties them genealogically to the ʻōiwi community, as the creation of all life (from plants to insects to human beings) is recounted in the epic story and weaved together in succession—just like a phylogeny in the field of genetics. Arthropods are also discussed in moʻolelo and kaʻao (two types of storytelling) (Paglinagwan, 2022). How these arthropods are described varies from revered cultural beings, such as having the designation of ‘aumakua (guardian), to agricultural pests. Therefore, there are many layers of traditional knowledge associated with arthropods, thus imbuing them with kinship.
Many of the arthropods in the Kumulipo were present in our data set. Therefore, sensitivity around the data from these Orders is elevated. However, since these orders represent dozens of families and species of arthropods, further discussion is required to untangle if each species is treated in the same way. Consequently, in the instance of the ant, it is a known introduced arthropod with significant negative impacts on ecosystems. How do you reconcile its presence in the Kumulipo with taxonomic origin and impact? These questions must be addressed by relevant cultural leaders, which we describe further in section 2.11.3 below.
2.11.2 Operationalizing and embedding Indigenous Data Sovereignty
As with the CARE principles, Paoakalani offers generalized, theoretical models for research governance and conduct in partnership with Indigenous communities. However, the research team is responsible for appropriately operationalizing these guidelines, specifically in the context of their research project. To operationalize the wishes of Paoakalani, our research team sought innovative modalities to safeguard IDS across all samples collected and data generated. For this, we utilized The Biocultural (BC) and Traditional Knowledge (TK) Notices developed by Local Contexts, an Indigenous-led organization, that are designed to provide Indigenous context and agency over Indigenous resources (Anderson & Christen, 2019). These Labels and Notices provide an extra-legal system of interest disclosure that creates space for community voices to be heard and address a pitfall in current Intellectual Property regimes that only recognize individual rights. To utilize the Label and Notices disclosure system, the project team created a researcher account through an online hub managed by Local Contexts. The application of these notices can be both visible as an icon in this publication (Figure 4) and as added fields in metadata tables with a specific project identifier (Liggins et al., 2021). However, commonly utilized genomic metadata standards still needed to be developed to include the disclosure of Indigenous rights, interests, and provenance information. Therefore, we added our own fields to the iBOL metadata manifest and filled them according to Mc Cartney, Anderson, et al. (2022); Mc Cartney, et al. (2023) using language from the Local Contexts Hub (Table 2).
2.11.3 Recognizing biocultural significance and considerations for Indigenous data futures
A limitation for our research team in using the BC and TK Notice is the lack of an account for the ʻōiwi community on the Local Context Hub. The usual streamlined process of using BC and TK notices is such that a community must have an account to be notified. From there, they decide how to address the research team and the data generated. In other Indigenous communities, community accounts are usually overseen by entities such as Tribal research review boards or designated oversight leaders. However, there is no centralized governance structure like this in Hawai'i for the ʻōiwi community. A typical path forward with this lack of governance structure is the formation of a hui (group) around central topics, which could be operationalized for managing genomic data among the ʻōiwi community. Previously, in the biological context, a hui comprised of ʻōiwi community members has been formed to discuss the cultural protocols, importance, and management of limu (seaweed; Kua ʻĀina Ulu ʻAuamo, n.d.), manu (birds; Paxton et al., 2022) and iʻa (fish; Vaughan & Caldwell, 2015).
- How did kūpuna (elders, ancestors) manage access to knowledge and resources?
-
Who should have access to project-generated arthropod data?
- Lineal descendants of the different ahupuaʻa (land division on a local scale) or moku (land division on a regional scale) sampled in Kona?
- Those who are kamaʻāina (familiar) and have pilina (connection) with the different arthropods?
- Other farmers and researchers looking to do work that continues to support an agricultural future in Kona?
The result of this would be a comprehensive and streamlined access approach that can be applied at varying scales, including on the Local Context Hub.
2.11.4 Establishing sustained and culturally appropriate Indigenous resource storage solutions
The process of forming a hui and engaging in needed conversations and decision-making takes a considerable amount of time. Although our project integrated a mechanism to support the disclosure of the Indigenous rights and interests associated with the species samples and sequencing information generated, to fully realize IDS, a solution was needed for where genetic resources and sequencing data would be stored. Here, in this project, long-term access to the community participants and long-term storage capacity was prioritized. Similar to many Indigenous communities worldwide, Hawai'i does not have an Indigenous-led or driven biobank or storage facility to store Indigenous samples obtained, so an external repository for both the physical and digital Indigenous resources was required. When selecting an appropriate external repository, it was important that the entity could act as a safe harbor for the resources collected until the community could establish a local repository. The selection strategy considered whether the entity had cultural awareness and training in Indigenous resource management and had the necessary infrastructure to support community accessibility and governance over the resources.
Therefore, for this project, we chose the Native BioData Consortium (NBDC) as an external repository for storing genomic data. NBDC is a not-for-profit, Indigenous-driven organization situated on the lands of the Cheyenne River Sioux Tribe in South Dakota. NBDC acts as a ‘safe harbor’ for Indigenous genetics resources and the associated sequencing data until a community has its own infrastructure to provide expertise, physical data storage, and legal services. It is also the only Indigenous-led bio-consortium in the United States. Both the raw and processed data, including the metadata, for this project will be hosted and stored on the NBDC server, with sequencing data access granted upon request to the community advisory board. Data decisions will be made on a case-by-case basis.
3 RESEARCH COMMUNICATION AND DISSEMINATION
Although project questions were not co-created with farmers, we evaluated whether project-generated data could address farmer interests and provide information on potential benefits to the farmers. We also considered how farmer interests align with our main research question and in the communication of our results. Importantly, in interpreting the results of our analysis, we show that data can serve many purposes and be communicated in different ways depending on the audience. In addressing these questions, we place a particular emphasis on the taxonomic origin of species, whether they are native or introduced, for a few key reasons: first, native taxa are often indicators of environmental change (Gillespie et al., 2008; Medeiros et al., 2013); second, native taxa are culturally significant and lastly, a whole suite of introduced taxa has caused considerable harm to ecological communities (Howarth, 1985) with significant consequences to Indigenous communities, such as the loss of culturally significant staple crops.
3.1 Overview of the data
In total, 222 OTUs were collected among all sites, of which 183 OTUs had introduced taxonomic status while 39 had native status. The orders Araneae, Coleoptera, and Lepidoptera represented many native OTUs (Figure S1). Conversely, most introduced OTUs held an equal proportion with some increase in Araneae and Coleoptera.
3.2 Exploring farmer interests
During conversations with farmers, we encountered two main interests regarding arthropods on their farms. Farmers asked: first, what arthropods are present on my farm? Second, how does this compare to other farms in the region? Some farmers also had specific questions about the Coffee Borer Beetle (‘CBB’; Hypothenemus hampei), which is plaguing their farms (Aristizábal et al., 2016). We could address the first and second questions with the data we generated from this project (Figure 4a–e). However, we did not detect any CBB despite the presence of beetles from the same family (Curculionidae) as CBB in our samples. The lack of CBB is potentially due to seasonality and sampling methodology. CBB tend to be more abundant with the development of cherries and collected by extracting them from cherries or beetle-specific traps (Follett et al., 2016). Therefore, the data could not address any CBB-related questions.
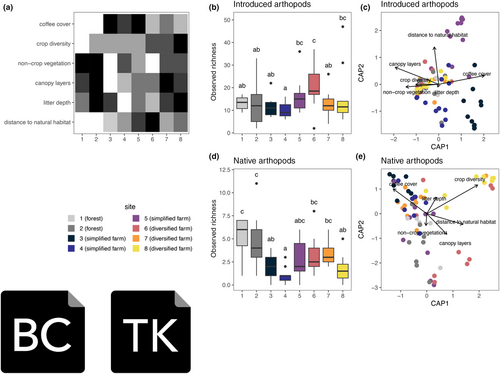
To address the farmer's questions, we analyzed the observed richness of both introduced and native arthropods at the site level (1–8), with forest sites included as a reference baseline. We examined environmental and management attributes known in other studies to alter the composition of arthropods across individual sites (Table 1). All sites varied in environmental and management attributes, which included measurements on on-farm vegetation (e.g. coffee cover, crop diversity, and non-crops), canopy structure, the amount of leaf litter on the ground, and the distance to natural habitat (Figure 4a). The variation among sites captures that agricultural land use is heterogenous, and seemingly similar sites (e.g. a few numbers of crops) can still have different habitat/structural properties (Benton et al., 2003).
A Tukey HSD showed introduced arthropod richness did vary among some sites, with some farm sites significantly more (e.g. site 6 vs. sites 1–4 and 7) or less (e.g. site 4 vs. sites 5, 6 and 8) than other sites (Figure 4b). In contrast, native arthropod richness appears to be more variable between sites. The forest sites had the highest number of native arthropods. However, diversified farm sites 6 and 7 were similar to the forest sites in richness. Curiously, simplified farm site 5 appears to have a subset of native arthropods on all sites. Diversified farm site 8 had lower observed richness than other diversified sites, with richness on par with simplified sites 3 and 4. This could be described by the landscape surrounding site 8 being dominated by monocultures, while the other sites had a more complex landscape mosaic. An additional factor could be the time in management, which we attempted to control by ensuring each farm had been using simplified or diversified management for at least five years. However, site 8 had not been in diversified management for as long as the other diversified sites.
Changes in arthropod richness can be attributed to some on-farm environmental and management properties. Coffee cover (p = .013) and distance to natural habitat (p < .001) significantly altered the richness (Table S1) and composition of introduced arthropods (Figure 4d). Site 3, a simplified farm with the highest proportion of on-farm coffee cover, had an observed introduced arthropod richness comparable to several other sites (Figure 4b). However, the arthropod community sampled from site 3 differed from other sites (Figure 4c). For native arthropods, richness was impacted by several site features, negatively by greater crop diversity and positively with more canopy layers, but proximity to natural habitat had no effect. Interestingly, diversified farm site 8, which had the highest crop diversity, had one of the lowest numbers of native arthropods (Figure 4d); yet, the composition of this arthropod community was distinct from other farm sites due to its high level of crop diversity, especially Polynesian crops (see below in section 3.3 for further discussion; Figure 4e). Therefore, there is a potential for farms, even those located within an area with little natural habitat, to support a unique composition of arthropods through on-farm management practices such as increasing crop diversification and including more Polynesian crops.
A SIMPER analysis identified the native and introduced OTUs contributing the most to the differences between sites (Figure 5a,b). For introduced arthropods, some of the most important OTUs driving community variation included OTU9 (Entomobryidae, 10.2% variation), OTU3 (Amphipoda, 6.89% variation), OTU15 (Brachymyrmex cordmoyi, 4.52% variation) and OTU7 (Entomobryidae, 4.37% variation) (Figure 5a). OTU9 and OTU7 belong to the springtail family, which are detritivores that thrive in soil, and are some of the most abundant introduced OTUs across all sites. Springtails may be particularly abundant in diversified farm sites due to management strategies that promote the build-up of leaf litter and the use of mulch. Curiously, OTU15, an ant known to thrive in the Neotropics, is mainly abundant in simplified farm sites.
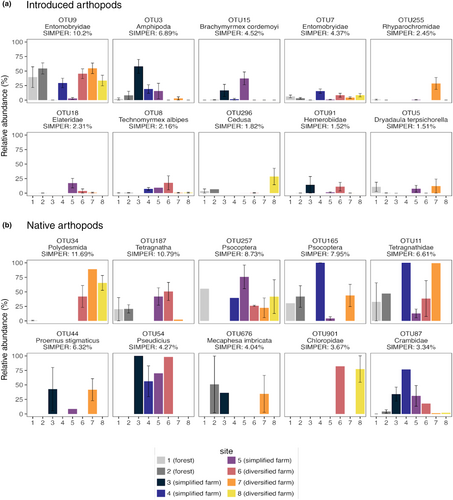
In terms of native arthropods, some of the most important OTUs driving community variation include OTU134 (Polydesmida, 11.69%) OTU187 (Tetragnatha, 10.79%), OTU257 (Psocoptera, 8.73%) OTU165 (Psocoptera, 7.95%) and OTU11 (Tetragnathidae, 6.61%) (Figure 5b). A commonality among all of these OTUs is that they have generalist feeding habits. OTU34 is a detritivore in the millipede family and is abundant at diversified farm sites. Again, this may be explained by the soil and leaf litter enhancement strategies on these diversified farms compared to simplified ones. OTU11 and 187 belong to Tetragnathidae, a well-studied family of spiders in Hawai'i that feed on various prey. These OTUs are well represented in farm and forest sites. Pscoptera (OTU257 and OTU165) were abundant across all sites and feed on lichen, fungi, and plant materials on various plant species. We further explore hypotheses on what mechanisms may be behind the retention of certain native taxa in farm sites below (see Section 3.3).
Drivers of site-by-site differences in arthropod richness and composition appear ambiguous, which could be aided by including more farmer participants/sites in future sampling and, thus, increasing statistical power. Yet, the data we collected could be more meaningful to individual farmers if it is not just aggregated to examine patterns that drive the number or composition of arthropods. Therefore, our farmer communication plan involves sharing a flier with individualized information for each farmer, including comprehensive information on the arthropod community detected on their farm. For example, by providing the trophic assignments for nearly all taxa observed on their farm, farmers can use this information to match what they see on the farm with the species list and hone in on pest species they may be encountering. Then, farmers can decide if they wish to engage in forms of Integrative Pest Management or work with the University of Hawai'i Extension or Natural Resource Defence Council to further inquire about the benefits of the species present (i.e. conservation payment programs). After receiving the flier, if a farmer wishes to engage further and learn more, we invite them to attend online or in-person one-on-one or group meetings. A communication plan ensures that project data will make it back to the community meaningfully, which is sometimes in contrast to project goals. This dual approach allows farmers to engage based on their comfort and interest.
3.3 Addressing our ecological question
After addressing farmer interests, the research team sought to understand our main research question to understand how the degree of agricultural diversification alters the diversity and composition of arthropod communities across sites with distinct distances to natural habitats. There is an increasing understanding that the diversification practices within agroecosystems, rather than just the presence of agriculture, play an instrumental role in local and landscape biodiversity patterns (Esquivel et al., 2021). Yet, diversification practices are heterogeneous––as demonstrated in the above-mentioned site-by-site variation in agricultural practices (Figure 4B). Therefore, we created an agricultural diversification index to assess how the culmination of practices (rather than a singular feature) impacts the diversity and composition of arthropods.
We expected a general positive effect of agricultural diversification on both introduced and native arthropods that would be magnified when sites were closer in distance to natural habitat. Yet, we only partially observed these trends. The observed richness of introduced and native arthropods increased with agricultural diversification (Figure 6a,c). Surprisingly, however, when sites were further from natural habitat, native arthropod richness decreased on more diversified sites (Figure 6c). A possible explanation could be that the native species are predated on or in competition with the high amount of introduced species in these systems. This high amount of introduced species is spread across a diversity of orders (Figure S1A) representing various trophic positions, of which orders such as Coleoptera and Araneae, which commonly hold predator trophic positions, are especially abundant.
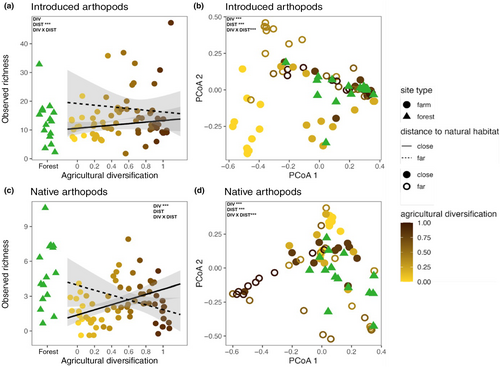
We expected a combination of diversification practices, such as the number of different crops and non-crops, could create habitat/opportunities for native arthropods (Figure 6e). With the shorter distance between farm sites and natural habitat, there is potential for repeated colonization from the natural habitat to the farm (i.e. a spillover effect), especially on farm sites with high agricultural diversification. As a result, the combination of agricultural diversification and proximity habitat may reduce competition between native and introduced arthropods. Further, more simplified farm sites also had a greater richness of native arthropods than diversified sites at similar distances. One possible explanation is that the more diversified farm sites were dominated by introduced arthropods (Figure 6a; Figure S1A). This suggests agricultural diversification, especially on sites further from natural habitat, may present opportunities for new, introduced species to establish and, consequently, may increase competition with native arthropods. In contrast, more simplified farms may generally have less habitat for introduced arthropods, thus presenting a less competitive environment for native species.
Despite the reduction in native arthropod richness, the further diversified farm sites harbored a unique composition of introduced and native species (Figure 6b,d). The composition of the native species present was heavily represented by mobile arthropods that feed on plant material or detritus (in the orders Lepidoptera, Diptera and Psocoptera; Figure S1B; Figure 6b). In addition, one particular group of arthropods in the families Crambidae and Chloropidae, known to feed on Polynesian crops, was highly abundant on the furthest site dominated by Polynesian crops (e.g. Maiʻa (banana; Musa acuminata) and kō (sugarcane; Saccharum officinarum) (Swezey, O.H., 1954)).
Unsurprisingly, the forest sites had the highest native richness (Figures 4c and 6c ). However, few species were exclusively found in these sites, as there was much taxonomic overlap with farm sites (Figure 6e). The subset of arthropod species only present on these forest sites belongs to orders with narrower ranges because they co-evolved with specific plant taxa such as Hemiptera (Roderick & Metz, 1997; Figure S1B; Figure 5b). The general lack of unique taxa is likely attributed to lower-elevation forest sites being inundated with introduced flora and fauna. Both forest sites had invasive flora that spread fast, including Yellow Ginger (Hedychium flavescens) and Mickey Mouse plant (Ochna serrulata). Taken together with the Polynesian crop results above, there is room for new management paradigms.
Preserving and restoring native flora in these lowland forests is often labor-intensive and expensive due to the invasive traits of many introduced flora, often leading to forest systems remaining degraded. To combat this trend, hybrid approaches of restoration utilizing Polynesian and non-invasive crops alongside native plants as tools have been proposed (Burnett et al., 2006; Ostertag et al., 2020; Winter et al., 2020), especially as a means to connect and prompt the access of ʻōiwi community members to their lands (Hutchins & Feldman, 2021; Lincoln et al., 2018). Still, many in the conservation field believe native arthropods cannot be found on agricultural sites. However, our findings support the potential of hybrid systems utilizing Polynesian and other crop species to support native arthropod biodiversity.
4 CONCLUSION
This paper described the tangible steps we took to operationalize IDS to use genomic data in the Kona Field System on Hawai'i Island. We recognize there is continued room for improvement and engagement at each step of our workflow. Future work should include more co-designing with community members from the outset. In terms of the ecological portion of this paper, since the study utilized limited pilot data, future sample collection should be more robust to adequately address ecological questions, such as measuring landscape heterogeneity and explicitly conducting a study to look at a farm management chronosequence.
- Take some time to reflect on your motivation to study a particular system or work with a community. Will your work detract from others in the community already conducting similar work? Is there a way to empower or build a partnership alongside that work? Engage in critical conversations with the project team on intention before and during the project. Understand your positionality to the land and community you seek to work with matters.
- Seek the resources to understand the history of the communities you seek to work with and how to appropriately engage (or not). Native-land.ca is a tremendous web resource for determining the native lands on which your research takes place. In the case of the United States, Tribes often have a website with the appropriate contact information for a research board or Tribal council. In addition, several universities have a tribal liaison who works to bridge the university with local Indigenous communities. In terms of communities with no centralized governance structure, as was the case in this paper, there are often local non-profit and government organizations that can offer guidance, such as a natural resources department or community health organization.
- Be open to having critical conversations and receiving feedback from community members and partners. You may be unable to conduct the specific project components you intended. Again, your positionality matters.
- Allocate an adequate amount of time to establish a connection with a community. The timeline to achieve all components varies widely. It depends on the context of your positionality, the community you seek to engage with and the nature of your research. The first step in establishing a connection with a community should be done respectfully and provided a sufficient amount of time. Establishing a meaningful relationship with a community can take years in it itself. Beginning engagement means you are open to sustaining a long-term relationship.
AUTHORS' CONTRIBUTIONS
LH and RG conceptualized the arthropod study. LH and AG collected the arthropod samples. LH and NG performed molecular processing of the samples. AG, NG, and LH conducted data analysis. AM and LH conceptualized the data sovereignty framework. LH and AM wrote the manuscript with input and comments from all co-authors.
ACKNOWLEDGEMENTS
We thank the farmers and community members that welcomed us onto their lands and shared space with us. We thank Isabel Lee-Park, Juliet Capriola, and Victoria Chen for their help in processing arthropod samples for DNA extraction. We thank Kevin Chang, Natalie Kurashima, and members of the Local Contexts hub for insightful conversations that helped to shape our data sovereignty workflow. We thank Cynthia King (Department of Land and Natural Resources) and Leah Laramee (Natural Area Reserve System) for access to state forest lands. Finally, we thank the reviewers for their insightful and helpful comments that greatly improved this manuscript, LH was supported by a Berkeley Food Institute seed grant and a National Science Foundation INFEWS fellowship.
CONFLICT OF INTEREST STATEMENT
All authors declare no conflicts of interest.
BENEFIT-SHARING STATEMENT
This article has benefitted from the input of several Indigenous communities and is intended to support greater benefit-sharing consistent with the FAIR and CARE data principles. Use of the Local Contexts Notices ensures appropriate acknowledgement and recognition of the communities that have contributed to the project. The Biocultural Notice for this project (UPID: https://localcontextshub.org/researchers/projects/33) discloses cultural rights and responsibilities that need further attention for any future sharing and use of this material or data. This Notice recognizes the rights of Indigenous Peoples to permission the use of information, collections, data and digital sequence information (DSI) generated from the biodiversity or genetic resources associated with traditional lands, waters and territories.
Open Research
DATA AVAILABILITY STATEMENT
All sequencing data generated by the project have been/will be archived in the Native Biodata Consortium repository that will act as a safe-harbor of the data until local capacity can be built. To obtain access, please send your requests to [email protected]. Requests should include name, affiliation and funders of the research team, as well as a one-page outline of how the data will be utilized if access is granted and how this use will benefit the community and society at large. Appropriate metadata associated with the sequencing data is available through BOLD (Code: DS-HULI), noting that all culturally salient metadata has been redacted consistent with Darwin Core.




