Assessing the utility of whole genome amplified DNA for next-generation molecular ecology
Abstract
DNA quantity can be a hindrance in ecological and evolutionary research programmes due to a range of factors including endangered status of target organisms, available tissue type, and the impact of field conditions on preservation methods. A potential solution to low-quantity DNA lies in whole genome amplification (WGA) techniques that can substantially increase DNA yield. To date, few studies have rigorously examined sequence bias that might result from WGA and next-generation sequencing of nonmodel taxa. To address this knowledge deficit, we use multiple displacement amplification (MDA) and double-digest RAD sequencing on the grey mouse lemur (Microcebus murinus) to quantify bias in genome coverage and SNP calls when compared to raw genomic DNA (gDNA). We focus our efforts in providing baseline estimates of potential bias by following manufacturer's recommendations for starting DNA quantities (>100 ng). Our results are strongly suggestive that MDA enrichment does not introduce systematic bias to genome characterization. SNP calling between samples when genotyping both de-novo and with a reference genome are highly congruent (>98%) when specifying a minimum threshold of 20X stack depth to call genotypes. Relative genome coverage is also similar between MDA and gDNA, and allelic dropout is not observed. SNP concordance varies based on coverage threshold, with 95% concordance reached at ~12X coverage genotyping de-novo and ~7X coverage genotyping with the reference genome. These results suggest that MDA may be a suitable solution for next-generation molecular ecological studies when DNA quantity would otherwise be a limiting factor.
Introduction
As the cost of both second- and third-generation DNA sequencing continues to plummet (Glenn 2011), ecological genomic studies will be feasible for the majority of individual laboratories, even those operating on a modest budget. Indeed, the reduced costs of modern sequencing technologies are enabling a more cost-efficient approach to empirical population genetic research than traditional programmes that, for example, utilized molecular cloning procedures for marker development. For example, one run on an Illumina MiSeq results in thousands of potentially suitable microsatellite markers to study rates and patterns of gene flow in nonmodel species (Castoe et al. 2012). These technological advances are now able to provide exponentially more data at a reduced cost per base pair vs. traditional Sanger sequencing-based methods (Glenn 2011).
Although next-generation DNA sequencing (NGS) has found a home in nearly all biological disciplines, these methods rely an initial library preparation step that requires a relatively large quantity of starting material for adequate construction (Quail et al. 2012). For example, a library insert size of 10 kb for sequencing on a PacBio RS II currently requires approximately 10 μg of starting gDNA (Pacific Biosciences). Although DNA libraries can be constructed with substantially less DNA for sequencing on other platforms using alternative techniques, DNA quantity can nonetheless be a limiting factor for genomic workflows. This issue has been addressed numerous times in the field of clinical and forensic research (see Lovmar & Syvänen 2006 for review).
Obtaining high concentrations of whole genomic DNA (gDNA) from large quantities of fresh tissue samples for NGS is certainly feasible in many cases. However, when assessing natural populations of rare species researchers are often forced to work in suboptimal conditions that include inadequate preservation methods, limited sampling regimes, and suboptimal tissue type and quantity. Molecular ecological studies, for example, might rely on noninvasive tissue sampling methods to obtain genetic material that often exhibits substantially reduced DNA quantity and perhaps quality (Taberlet & Luikart 1999; Taberlet et al. 1999; Waits & Paetkau 2005). In addition, old tissue samples preserved in inadequate buffer may result in reduced DNA yield that is of high molecular weight and generally yields biased sequencing data unfit for analysis (Taberlet & Luikart 1999; Taberlet et al. 1999). Further, ancient DNA and environmental DNA (eDNA) studies are often plagued by low-quantity, highly degraded DNA that puts limits both on the choice of sequencing procedures and more importantly the hypotheses that can be tested (Hofreiter et al. 2001; Pääbo et al. 2004; Taberlet et al. 2012b).
Although relatively unexplored in ecological and non-human evolutionary research, whole genome amplification (WGA) may be a viable method to substantially increase starting DNA yield for subsequent genetic and genomic analysis (see Lasken 2009 for review). In noninvasive strategies such as hair collection (particularly if samples are collected soon after they are shed from the animal), DNA is often of high molecular weight though in low copy number quantities. WGA may therefore be useful to increase DNA concentrations so that NGS can be performed. For ancient DNA and eDNA applications, WGA could complement previously existing laboratory methods (e.g. Rohland & Hofreiter 2007) and be useful for increasing DNA yield based on the limited number of target fragments long enough to be successfully amplified. Several different WGA methods and kits are currently available including degenerate oligonucleotide primed PCR (DOP; Telenius et al. 1992; Cheung & Nelson 1996), primer extension PCR (PEP; Zhang et al. 1992) and multiple displacement amplification (MDA; Dean et al. 2002). Unlike PCR methods that use Taq DNA polymerase and may result in amplification bias and highly skewed genome coverage (Dean et al. 2002; Lasken 2009), MDA techniques use a high-fidelity φ29 polymerase and random hexamers to provide a more stable amplification with a more uniform coverage and longer fragment lengths >10 kb (Dean et al. 2002; Hosono et al. 2003; Lovmar et al. 2003; Paez et al. 2004; Park et al. 2005; Pinard et al. 2006).
Although the use of MDA for increasing copy number may seem like a panacea when working with low-quantity DNA samples, amplification bias remains a concern when using these methods. Several studies have quantified potential bias in MDA, although the majority have either focused on few loci (e.g. Cheung & Nelson 1996; Dean et al. 2002; Hosono et al. 2003; Lovmar et al. 2003), genomic resources rarely available for nonmodel taxa (e.g. Barker et al. 2004; Paez et al. 2004), or relatively simple genomes (e.g. Abulencia et al. 2006; Pinard et al. 2006). Further, only a handful of studies have examined how bias scales with the use of the high-throughput DNA sequencing methods now common in both clinical and nonclinical research programmes (e.g. Pinard et al. 2006; ElSharawy et al. 2012). Thus, additional studies are needed to examine the efficacy of ‘next-generation MDA’, particularly for research applications that target thousands of loci for hundreds of individuals as is commonly observed in NGS applications in molecular ecology.
Genotyping-by-sequencing (GBS) methods such as restriction site-associated DNA sequencing (RADseq; Baird et al. 2008) and double-digest RADseq (ddRADseq; Peterson et al. 2012) have become popular approaches for interrogating thousands of genomic regions for up to hundreds of individuals at a modest sequencing cost (Davey & Blaxter 2010; Rowe et al. 2011; Narum et al. 2013). In general, these techniques are ideal for population genomic- and shallow-scale phylogenomic studies (e.g. Emerson et al. 2010; Hohenlohe et al. 2010, 2011, 2013; Peterson et al. 2012; Rubin et al. 2012; Eaton & Ree 2013; Nadeau et al. 2013; Wagner et al. 2013; Mastretta-Yanes et al. 2015; Schield et al. 2015). New analytical software packages such as Stacks (Catchen et al. 2011, 2013) and pyrad (Eaton & Ree 2013) are attractive in that they allow researchers to control multiple parameters to cluster sequences based on a priori knowledge about their study system (Davey et al. 2013). This flexibility helps solve some of the challenges for many of these methods, including adequately merging orthologous sequences within and among individuals while minimizing the clustering of paralogous markers. However, no study to date has tested for bias when performing ddRADseq on MDA samples. We focus our efforts on ddRADseq, as the method is extremely flexible (Peterson et al. 2012) and likely to become routinely used in ecological and evolutionary genomics for the foreseeable future.
Here, we examine genetic material from the grey mouse lemur, Microcebus murinus, to investigate the potential bias introduced with MDA and ddRADseq. By performing ddRADseq on the same set of individuals in duplicate – that is., those from both MDA and high quality gDNA – we test the null hypothesis that there are no observable differences in both relative genome coverage as well as inferred SNPs within individuals among sample types. We also estimate the potential for increased homozygosity in MDA samples due to allelic dropout. We compare our findings when genotyping de-novo vs. genotyping from mapping RAD sequences to an available mouse lemur reference genome. We find that sequence bias resulting from MDA is very low to nonexistent, especially when sequencing coverage is sufficiently deep. We thus hope that our results will find use for a wide variety of applications for studying molecular ecological and evolutionary processes in natural populations.
Methods
Samples and laboratory methods
We obtained fresh tissue (liver) samples from five Microcebus murinus individuals (MM1812, MM1842, MM1895, MM7011, MM7020) from the Duke Lemur Center (DLC, Durham, NC, USA) that were immediately preserved at −80 °C. We used the Qiagen DNeasy Blood & Tissue DNA Extraction Kit (Qiagen Sciences, Inc.) to obtain purified gDNA for each sample. All DNA extracts were treated with Qiagen RNAse A to remove any RNA contamination. Following purification and quantification on an Agilent Nanodrop 2000 spectrophotometer, MDA reactions were performed for each individual. For our study, we utilized the MDA technique used in the Qiagen REPLI-g Mini Kit following manufacturer recommendations (>100 ng of high molecular weight gDNA per reaction). Although we could have performed MDA reactions with substantially less starting DNA (to more closely mimic potential field conditions), we chose to follow manufacturer guidelines for a ‘best case scenario’ comparison. Our primary justification for this approach is that before potential bias with MDA and ddRADseq can be assessed when starting with exceptionally low DNA quantity (generally <1 ng), baseline studies are needed to test whether there is bias in the MDA reaction itself when used in conjunction with ddRADseq. Also of note is that total DNA yield from MDA is highly uniform irrespective of the quantity of starting material (Dean et al. 2002), and some studies have suggested that MDA efficiency may only be compromised at extremely low DNA input (see 4).
All MDA reactions were incubated overnight at 30 °C and subsequently heat-killed at 65 °C for 3 min. Products were run on a 2% agarose gel stained with ethidium bromide to check the quality of reactions. Quantification of unpurified MDA reactions on an Agilent Nanodrop 2000 yielded >300 ng/μL DNA per sample. To remove unincorporated nucleotides and clean MDA products, we used the Zymo Genomic DNA Clean & Concentrator Kit (Zymo Research) following specified protocols. Quantification of purified MDA samples yielded concentrations between 50 and 100 ng/μL. Although unpurified MDA reactions can be used for subsequent assays (Hosono et al. 2003), we purified our samples to minimize any potential bias introduced by unincorporated reagents.
To create ddRAD libraries, we followed the protocol of Peterson et al. (2012). The ddRADseq protocol was selected over single-digest RADseq for several reasons; (i) digesting with two restriction enzymes potentially eliminates the need for the expensive and error-prone step of randomly shearing the DNA; (ii) the efficiency of random shearing and subsequent library preparation steps will be more sensitive to both input DNA quantity and quality; (iii) restriction fragment length bias is a greater concern with RADseq (Davey et al. 2013); (iv) the majority of laboratories will have the necessary equipment to perform both MDA and ddRADseq in-house. Briefly, we first double digested the gDNA and MDA samples with the enzymes SphI and MluCI (New England Biolabs). Digestions (50 μL) were performed for 3 h at 37° C using ~1 μg DNA using the following master mix: 5 μL NEB buffer 4, 1 μL of each enzyme, 15–20 μL gDNA or MDA, and ddH20 to volume. Digests were then cleaned using 1.5X volume Agencourt Ampure Beads (Beckman Coulter) following manufacturer protocols. Ligation reactions (40 μL) were then performed to attach adapters (barcoded P1, P2) containing PCR and sequencing regions to each individual. Adapters were first diluted (from a 40 μm stock) targeting a value of fivefold excess adapters to complementary sticky ends following Peterson et al. (2012). The P1 adapter was diluted to a final working concentration of 0.406 μm, and P2 was diluted to 0.876 μm. Each reaction contained 4 μL T4 buffer, 1 μL T4 DNA ligase, 2 μL of each adapter, 50 ng DNA and ddH20 to volume. Samples were then incubated following Peterson et al. (2012). Ligated products were then combined, cleaned using Ampure beads and size-selected using a 2% agarose gel with a target range of 400–600 bp. The cleaned product was aliquoted into five separate PCR tubes and amplified using the Phusion High-Fidelity PCR Kit (New England Biolabs). All reactions contained the following mastermix (20 μL per sample): 4.4 μL ddH20, 4 μL 5X Phusion PCR buffer, 0.4 μL 10 mm dNTPs, 1 μL 10 μm of each primer, 0.2 μL DNA polymerase and 9 μL purified size-selected DNA. Cycling conditions used were as follows: denaturation at 98 °C for 30 s, 12X cycles of 98 °C for 10 s, 68 °C for 30 s, 72 °C for 30 s and a final extension step of 72 °C for 7 min. PCR products were pooled, cleaned with Ampure beads and eluted in EB buffer to a final volume of 40 μL. DNA was then quantified on a Nanodrop and Bioanalyzer 2100 (Agilent) and further size-selected using a Pippin Prep (Sage Science) because of the relatively wide range of fragments recovered by gel excision. The resulting fragments (~320–520 bp) were visualized on a TapeStation 2200 (Agilent) and subsequently sequenced on an Illumina MiSeq DNA sequencer (150-bp paired-end) at the Duke University Institute for Genome Science & Policy Sequencing Facility.
To increase the number of comparisons between gDNA and MDA and to ascertain if results were due to laboratory or sequencing error, we performed a second experiment starting from the original liver samples and followed the same protocol as above. However, in the second experiment size selection was performed using the Pippin Prep only (i.e. no gel excision). Sequenced fragments in this run were ~275–500 bp in length (mean = 375 bp). All subsequent comparisons between gDNA and MDA were performed twice (i.e. one set per repetition of the experimental procedures above).
Data demultiplexing and quality filtering
To check the quality of the data, we first imported the paired-end reads into FastQC. This enabled us to quantify several characteristics of the data, including to what fragment length the reads should be trimmed for subsequent analysis. We then used the pipeline Stacks v 1.20 beta (Catchen et al. 2011, 2013) to process our RAD data. Stacks was developed specifically to deal with short-read data generated through next-generation sequencing methods and is ideal for gleaning population genomic statistics from ddRADseq studies. Although other programmes handle ddRADseq data and have different relative strengths (e.g. pyrad for deep phylogenetic studies and Genome Analysis Toolkit [GATK] for quality-aware SNP calling), Stacks is generally the currently recommended software package based on ease of use, features and performance (Davey et al. 2013). Samples were first demultiplexed and quality-filtered using the process_radtags program. Because the quality of the data was high through nearly the full length of the reads (150 bp), we only trimmed sequences to 145 bp for all subsequent analyses. Loci were then built both de-novo and with the available M. murinus reference genome.
De-novo analysis
We first used Stacks to build loci de-novo. The entire pipeline was run 10 times [i.e. once for each duplicate pair (MDA vs. gDNA) for both sequencing runs]. To minimize potential errors when making comparisons between gDNA and MDA samples, only single-read data were analysed. Thus, following execution of process_radtags, we combined the demultiplexed fastq files from read-1 with read-1 sequences that were stranded (singletons—rem file) into a single file (per individual) for analysis. All sequences have been deposited into the Sequence Read Archive (Table S1, Supporting information).
A new program in the Stacks pipeline, rxstacks, was recently developed to further quality filter data and correct SNP calls and excess haplotypes. At present, this program cannot be implemented using the streamlined pipelines denovo_map.pl and ref_map.pl. Therefore, following demultiplexing and filtering, each Stacks program was run independently using the following sequence: ustacks > cstacks > sstacks > rxstacks > cstacks > sstacks. The pipeline was executed using the following settings: minimum depth of coverage required to create a stack (-m) = 3; maximum distance allowed between stacks (-M) = 4; maximum distance allowed to align secondary reads (-N) = 6; –max_locus_stacks = 3; enable removal algorithm (-r) and deleveraging algorithm (-d); maximum number of mismatches allowed between loci when building the catalogue (-n) = 3. For rxstacks, we used the following settings: –conf_filter –conf_lim 0.25, –prune_haplo –model_type bounded –bound_high 0.1 –lnl_lim −10, –lnl_dist. These parameters were carefully chosen based on the fact that each of the 10 data sets consisted of pairs of the same individual. For example, for both -M and -n, we presumed that these values would be efficient to cluster orthologous loci and simultaneously minimize clustering of divergent paralogous markers. Parameters were also chosen based on recommendations from previous studies (Catchen et al. 2013). Following the execution of the pipeline, we implemented the populations program to generate output to compare the gDNA data to MDA for each paired individual. Because inferred SNPs and haplotypes were affected by depth of coverage, we adopted a relatively conservative approach and only kept loci with a minimum stack depth of 20X (-m in populations). All other default settings were used. We created a MySQL database to house Stacks output, and results were visualized on the Stacks webserver. All subsequent filtering and processing were performed in UNIX. Bias between MDA and gDNA samples was quantified at the SNP level for each individual. Note that throughout the study, we refer to tags or RAD-tags as the entire ddRAD sequence (145 bp), whereas a locus refers to a single SNP marker.
Reference analysis
In addition to creating loci and genotyping de-novo, we were interested in using the available M. murinus genome to assemble our RAD-tags and call SNPs. We obtained a copy of the M. murinus genome (assembled scaffolds) from the Ensembl Genome Browser (useast.ensembl.org/index.html). We then used Bowtie2 v2.1.0 (Langmead & Salzberg 2012) to map the quality-filtered RAD-tags to the reference. We ran initial analyses using the paired reads specifying a maximum fragment length of 600 bp. Default settings were used for all mapping. SAMtools v0.1.17 (Li et al. 2009) was used to convert the resulting SAM files to BAM files to ease data manipulation. Visualization of BAM files was implemented in the software Tablet v1.13.08.05 (Milne et al. 2013) for QC. We extensively browsed the mapping results for different scaffolds of the reference to assess the quality of alignment, in particular regarding the mate pairs. Results indicated a relatively large fraction of mates were stranded (i.e. one mate was unmapped or mapped to a different scaffold). This issue could be due to any number of potential factors including issues with the assembly of contigs and scaffolds, gene duplication, repetitive DNA, mutation, etc. To be conservative and to minimize potential errors mapping to the reference (which would subsequently influence genotyping), we used only the single-read data for alignment. Using the single-read data for mapping also enabled us to make a fair comparison to the results of the de-novo analysis. Therefore, we reran Bowtie2 for each individual using the single-read data only. Default settings were again used and the resulting alignment was post-processed in SAMtools and Tablet. We then reran Stacks an additional 10 times using the same settings as the de-novo analyses but substituting ustacks for pstacks. Catalogues were created using information both regarding genomic position of reads (-g) and the maximum number of nucleotide differences for consensus sequences (-n 3).
Finally, as read depth continues to be a major hurdle to accurately score genotypes from RAD-based data set (Davey et al. 2013), we were also interested in assessing how the minimum level of coverage for genotyping might influence SNP concordance between gDNA and MDA samples. Therefore, we ran additional populations analyses specifying the following minimum thresholds for coverage: 0X, 5X, 10X and 15X. These analyses were performed both de-novo and with the reference for sequencing Run-1 only. Output files from all Stacks analyses can be obtained from Dryad (doi:10.5061/dryad.83dc2).
Results
Quality filtering
We first assessed the quality of the paired RAD-tags using FastQC. Both sequencing runs were of high quality, with median per base sequence qualities (Phred score) ranging from 32 to 39 (majority 39). Median per base quality scores after removing adapter and barcode sequence (i.e. data analysed in Stacks) ranged from 37 to 39. Likewise, the vast majority of the sequence reads had a mean Phred score >38, indicating that the data were of high quality. The correct cut site was found in the majority of fragments for both restriction sites (CATG and AATT), indicating that the digestion and ligation proceeded as expected. The data were then imported into Stacks where samples were demultiplexed and filtered.
For Run-1, out of a total of 18 715 106 sequences, 96 214 (0.5%) had ambiguous barcodes, 196 448 (1%) were of low quality and 106 438 (0.6%) had an ambiguous RAD-tag. Removing these reads left 18 316 006 reads (~98%) for subsequent analysis. The mean number of reads for the gDNA samples was 1 915 548, whereas the mean number of reads for the MDA samples was 1 808 230. Run-2 yielded 30 205 860 total paired reads, 591 620 (2%) of which had ambiguous barcode drops, zero low-quality read drops and 244 581 (0.8%) ambiguous RAD-tag drops. Removing these reads left 29 369 659 reads (~97%) for subsequent analysis. The mean number of reads for the gDNA samples was 3 032 860, whereas the mean number of reads for the MDA samples was 2 889 988.
De-novo analysis
The de-novo analysis of RAD-tags for Run-1 yielded between 34 469 and 70 454 unique stacks per individual, with a total number of tags in each catalogue ranging between 51 299 and 80 250 (Table 1). The de-novo analysis for Run-2 yielded between 72 137 and 140 158 unique stacks per individual, with a total number of tags in each catalogue ranging between 127 648 and 166 585 (Table S2, Supporting information). There was no observable difference between gDNA and MDA samples for the number of tags and SNPs generated within individuals. After extensive filtering of the data at a conservative level (minimum of 20X coverage), the total number of tags in catalogues was greatly reduced in both Run-1 and Run-2 (Table 1; Table S2, Supporting information). We then used the remaining tags to examine individual shared SNP loci to compare rates of concordance between gDNA and MDA. Results suggested a very high level of concordance (~98%) among gDNA and MDA in both runs for the SNP loci that remained after filtering (Fig. 1a; Fig. S1a, Supporting information).
| Individual | Barcode | Source | Unique stacks | SNPs | Total number of tags in catalogue | Total number of retained tags at 20X |
|---|---|---|---|---|---|---|
| MM1812 | GCATG | gDNA | 53 630 | 7 776 350 | 71 280 | 8554 |
| CAACC | MDA | 64 456 | 9 346 120 | |||
| MM1842 | GGTTG | gDNA | 63 059 | 9 143 555 | 70 978 | 10 413 |
| CGATC | MDA | 58 475 | 8 478 875 | |||
| MM1895 | AAGGA | gDNA | 64 590 | 9 365 550 | 70 768 | 8988 |
| TCGAT | MDA | 54 785 | 7 943 825 | |||
| MM7011 | AGCTA | gDNA | 64 360 | 9 332 200 | 80 250 | 11 309 |
| TGCAT | MDA | 70 454 | 10 215 830 | |||
| MM7020 | AACCA | gDNA | 49 518 | 7 180 110 | 51 299 | 3679 |
| ACACA | MDA | 34 469 | 4 998 005 |

Reference analysis
We first separately mapped all single-read RAD-tags for each sample to the currently available M. murinus genome. Alignment rate was consistent both among samples and among runs at ~80%. Approximately 50% of RAD-tags aligned once to the genome, whereas ~30% aligned more than once and ~20% did not align. There was no significant difference in alignment rate between gDNA and MDA samples in either Run-1 (Wilcoxon V = 12, P = 0.3125) or Run-2 (Wilcoxon V = 15, P = 0.0625).
Results from the reference analysis were generally concordant with those of the de-novo pipeline. The number of unique stacks per individual in Run-1 ranged from 34 776 to 84 764 and the total number of tags in catalogues ranged from 45 438 to 74 073 (Table 2). The number of unique stacks per individual in Run-2 ranged from 81 781 to 164 391, and the total number of tags in catalogues ranged from 110 979 to 130 808 (Table S3, Supporting information). Like the de-novo analysis, statistics were similar across all gDNA and MDA samples, and again, the total number of RAD-tags was substantially higher in Run-2. Further, SNP concordance among duplicate gDNA and MDA samples was high for the shared SNP loci remaining after filtering at 20X coverage (~98%; Fig. 1b; Fig. S1b, Supporting information).
| Individual | Barcode | Source | Unique stacks | SNPs | Total number of tags in catalogue | Total number of retained tags at 20X |
|---|---|---|---|---|---|---|
| MM1812 | GCATG | gDNA | 58 314 | 8 455 530 | 66 093 | 5829 |
| CAACC | MDA | 77 056 | 11 173 120 | |||
| MM1842 | GGTTG | gDNA | 72 393 | 10 496 985 | 65 876 | 6847 |
| CGATC | MDA | 67 328 | 9 762 560 | |||
| MM1895 | AAGGA | gDNA | 74 690 | 10 830 050 | 64 998 | 5959 |
| TCGAT | MDA | 61 268 | 8 883 860 | |||
| MM7011 | AGCTA | gDNA | 73 815 | 10 703 175 | 74 073 | 7304 |
| TGCAT | MDA | 84 764 | 12 290 780 | |||
| MM7020 | AACCA | gDNA | 52 092 | 7 553 340 | 45 438 | 2984 |
| ACACA | MDA | 34 776 | 5 042 520 |
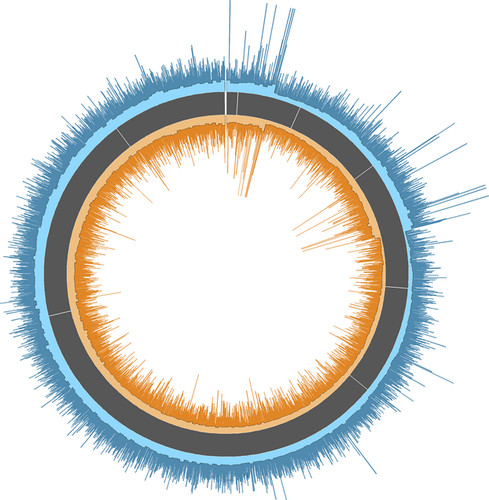
To further assess similarity in RAD-tag location and coverage depth among gDNA and MDA, a Circos (Krzywinski et al. 2009) diagram was generated to visually inspect coverage across the grey mouse lemur genome. We first performed two separate mapping analyses in Bowtie2 (all gDNA and all MDA for Run-1) and used the two resulting SAM files to generate the figure. These results confirmed the results of the SNP concordance assessments; both sample preparation techniques, gDNA and MDA, had consistent levels of average and maximum coverage depth across the M. murinus genome (Fig. 2).
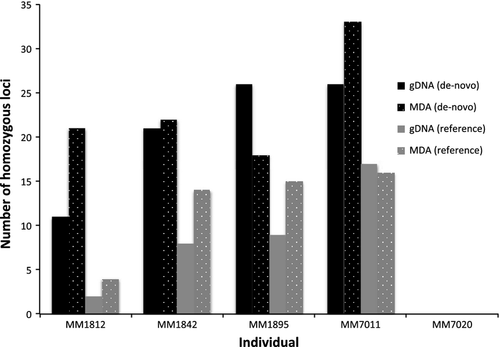
Allelic dropout
Previous studies have suggested that allelic dropout (i.e. failure to detect alleles that are actually present) may be a concern when working with wgaDNA (Lovmar et al. 2003; Handyside et al. 2004; Sun et al. 2005). For both pipelines (de-novo, reference) and sequencing runs (Run-1, Run-2), we calculated the number of shared SNP loci that were homozygous and heterozygous for both gDNA and MDA. For Run-1, there was no significant difference in the proportion of homozygotes among gDNA and MDA either genotyping de-novo (Wilcoxon V = 3, P = 0.5839; Fig. 3) or using the reference (Wilcoxon V = 1, P = 0.1975; Fig. 3). Similar results were obtained for Run-2 de-novo (Wilcoxon V = 8, P = 1; Fig. S2, Supporting information) and with the reference (Wilcoxon V = 0, P = 0.1003; Fig. S2, Supporting information), indicating that the MDA protocol had no appreciable effect on SNP calls and levels of homozygosity.
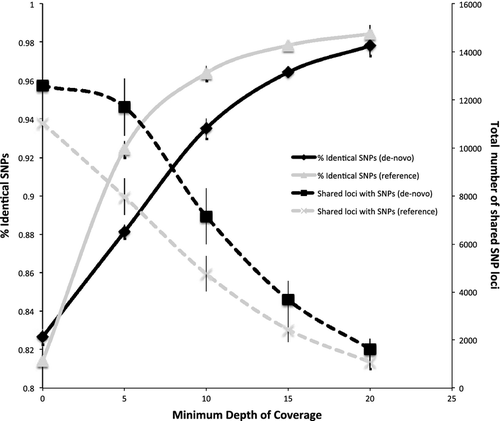
Coverage and SNP concordance
As recent studies have suggested that genotyping accuracy of RAD data is heavily reliant on levels of coverage (Davey et al. 2013), we tested the hypothesis that levels of SNP concordance among gDNA and MDA were related to the minimum level of coverage required to call genotypes. This minimum coverage level likely depends on whether the data are analysed with or without a reference genome. We therefore performed analysis both with and without the M. murinus reference. We tested the following minimum coverage thresholds: 0X, 5X, 10X, 15X and 20X. Results suggested that 95% SNP concordance among gDNA and MDA was obtained with a threshold of ~7X with the reference and ~12X with the de-novo assembly (Fig. 4). Concordance rates began to asymptote at about 5X coverage with the reference and at approximately 10X de-novo. At 20X coverage, there was no difference in gDNA and MDA SNP concordance among reference-based vs. de-novo genotyping. The average number of shared SNP loci across all individuals was consistently higher for de-novo analyses vs. reference analyses for a given sequencing depth out to about 15X coverage (Fig. 4).
Discussion
Advances in DNA sequencing technologies have revolutionized ecological and evolutionary studies of natural populations, enabling researchers to test a suite of hypotheses that were previously beyond the scope of sequencing technology. With these new methods come challenges ranging from adequate experimental design to data analysis. We focus our efforts here on quantifying bias in genome coverage and SNP genotypes by comparing duplicate gDNA and MDA samples from the grey mouse lemur, M. murinus, using ddRADseq. Although other WGA methods are available, we focus solely on MDA as this technique is generally considered to be one of the more reliable methods (Lasken 2009). We use ddRADseq as our method of choice for genetic characterization because of the increasing popularity of restriction enzyme-based methods in population genomic and shallow-scale phylogenetic studies (Emerson et al. 2010; Hohenlohe et al. 2010, 2011, 2013; Peterson et al. 2012; Rubin et al. 2012; Mastretta-Yanes et al. 2015; Schield et al. 2015). Our results suggest both adequate genome coverage and SNP concordance among multiple individuals and separate independent sequencing runs. Genotype mismatches between MDA and the control gDNA were rare in both replicates of the experiment, suggesting that MDA has no appreciable effect on ddRADseq experiments when using an acceptable level of input DNA (>100 ng). These results are congruent with previous studies that quantify bias in MDA using alternative methods (e.g. Dean et al. 2002; Hosono et al. 2003; Lovmar et al. 2003; Barker et al. 2004; Sun et al. 2005), For example, SNP and STR genotype concordance rates between MDA and gDNA have been estimated to range from 70% to 100% (majority >90%), depending on the methodology utilized (see Lovmar & Syvänen 2006 for review). However, many of these studies were based off a relatively low number of markers. Our results from ddRADseq are concordant with both previous and more recent studies that examine how bias scales with NGS data (e.g. Pinard et al. 2006; ElSharawy et al. 2012) and suggest that MDA may be an important tool for ecological and evolutionary genomics.
Ecological applications of MDA
A primary motivation for this study comes from our own experiences with suboptimal tissue preservative for long-term storage of rare samples from Madagascar. Further, given the highly endangered status of virtually all endemic Malagasy vertebrates, we must typically rely on hair and other sample types that have low amounts of endogenous DNA (Taberlet & Luikart 1999; Taberlet et al. 1999). Moreover, these samples must be initially preserved under field conditions, and due to U.S. regulatory oversight, must often be stored for several to many months at a time as they await exportation. In such cases, samples typically contain low-quantity (~3–10 ng/μL), high molecular weight DNA that is suitable for MDA (Dean et al. 2002). The finding of no substantial bias in MDA samples suggests that these tissue samples, which usually cost thousands to tens of thousands of dollars in time, equipment and labour, can be saved for later use with NGS methods and lead to larger sample sizes for population genomic and phylogeographic studies. It should be noted, however, that many WGA procedures, including MDA, may not produce a reliable and unbiased amplification when working with highly degraded template (Wang et al. 2004). As no study has assessed the potential biases of MDA and ddRADseq, the present study serves to establish a baseline operating within ideal conditions of >100 ng of high molecular weight starting DNA template (for further discussion see 4.2). To maximize the potential of MDA for noninvasive sampling regimes, tissue samples should be collected soon after they are released from the animal to minimize the quantity of degraded DNA in the sample. A simple DNA extraction followed by gel electrophoresis can be performed to assess whether the samples are good candidates for MDA.
Another potential use of WGA generally and MDA specifically lies in eDNA metabarcoding studies, which are increasing in popularity at an extraordinary rate. These studies use DNA present in environmental samples such as water or soil to detect the presence or absence of particular species (Ficetola et al. 2008; Taberlet et al. 2012a,b). Thus, eDNA can be classified as a second type of noninvasive DNA sampling and again, DNA quantity and quality can be a limiting factor depending on the amount of time elapsed between the time DNA molecules were shed to the time they were preserved (Dejean et al. 2011; Thomsen et al. 2012a,b; Goldberg et al. 2013; Barnes et al. 2014). It is also highly likely that eDNA molecules from different species are present in different concentrations, and it is presently unknown how this asymmetry may affect species detection and estimates of relative abundance. This may be particularly true for rare species, whose DNA may be represented in exceptionally low quantities (Boessenkool et al. 2012). MDA may be one solution to simultaneously amplify all DNA in an environmental sample prior to library construction, yielding more accurate species inventories. However, it may be possible that MDA introduces bias in eDNA applications by preferentially amplifying DNA fragments from species that have not passed a threshold of degradation. This potential trade-off is an area ripe for research.
Finally, MDA has shown promise in the field of metagenomics (Abulencia et al. 2006) and for DNA amplification in single bacterial cells (Raghunathan et al. 2005). Our results support the notion that MDA may be useful for population genomic studies of microbes to fully characterize genomic diversity and population structure of both bacteria and viruses. This is a particularly exiting avenue of research as more accurate estimates of genomic diversity for common human pathogens can be ascertained. Further, comprehensive genomic assays for these pathogens would enable more robust estimations of genomic patterns of adaptation and how these patterns correlate with particular demographics.
Considerations and limitations
Although our results report no substantial bias between gDNA and MDA using ddRADseq, there are several limitations of the current study that must be addressed. First, we base our conclusions on a single WGA method and kit (Qiagen REPLI-g). A variety of other MDA kits are presently available including Illustra GenomiPhi and TempliPhi (GE Healthcare Life Sciences). However, the principle and chemistry between these kits is virtually identical and is thus likely to provide similar results.
Second, allelic dropout in heterozygous individuals can be an issue when working with wgaDNA (Lovmar & Syvänen 2006). The probability of allelic dropout appears to a factor of both the method of WGA used and the quantity of starting material (Handyside et al. 2004; Bergen et al. 2005a; Sun et al. 2005). More specifically, PCR-based WGA methods or starting DNA quantities <1 ng may introduce some degree of both locus and allelic bias (Cheung & Nelson 1996; Dean et al. 2002; Sun et al. 2005). Results from additional studies suggest DNA input quantities between 3 and 100 ng per MDA reaction to maximize genome coverage and minimize allelic dropout and imbalance (Lovmar et al. 2003; Bergen et al. 2005b). We follow these guidelines in the present study by utilizing the MDA approach with starting DNA concentrations that are relatively large (>100 ng as per manufacturer recommendations). Indeed, our results suggest weak evidence for allelic dropout in MDA samples, as the proportion of homozygotes in shared loci is similar for both gDNA and MDA.
Third, although our results serve as a baseline estimate and suggest limited bias within MDA and ddRAD libraries when strictly following manufacturer guidelines, future studies should compare the concordance of gDNA and MDA among high (10–100 ng) and low-quantity (<10 ng) DNA template. These comparisons should be made within a single library and sequencing run to minimize any potential exogenous bias. For example, a suitable follow-up study would be to create libraries consisting of both high quantity gDNA from blood and low-quantity DNA from hair (to be amplified by MDA), with both sample types originating from the same animal. This combined ‘testing the kit + ‘testing the sample’ approach will highlight more of the potential limitations when using MDA in ecological and evolutionary research.
Fourth, we focus our efforts solely on GBS techniques (i.e. ddRADseq) as these are becoming methods of choice for next-generation population genomic and phylogeographic research (Davey & Blaxter 2010; Rowe et al. 2011; Narum et al. 2013). Additional studies are needed to determine how alternative NGS approaches commonly used for similar ecological applications such as sequence capture methods compare with our results. Further, we test for bias only using ddRADseq while bias may prove more substantial in alternative RAD-based protocols, particularly those that involve a random shearing step. However, we anticipate that results will be highly congruent as evidenced from other recent studies quantifying bias in MDA with NGS (Pinard et al. 2006; ElSharawy et al. 2012). Along similar lines, we use a single software package, Stacks, to analyse our data. We focus on Stacks because the software was specifically designed to process RAD-based markers and is the generally accepted analytical method (Davey et al. 2013).
Fifth, we use a relatively low-quality reference genome (~2X) in our analysis, which may reduce mapping efficiency. This may suggest why at any given coverage depth, the number of shared loci among samples is greater when genotyping de-novo than with the reference (Fig. 4). Interestingly, however, results regarding the efficiency of MDA are virtually identical in both cases, suggesting that the use of a low-quality reference is not a hindrance to our conclusions.
Our study design and objectives allow us to test how genotyping efficiency may be related to minimum depth of coverage. Compared to fragment analysis, genotyping NGS data are far less straightforward with multiple factors that must be considered and incorporated for accurate calls (Davey et al. 2013). As more NGS data are collected, new genotyping programs will continue to be developed and the strengths and weaknesses of methods will have to be compared. Based on the maximum-likelihood method in Stacks, our results suggest that a minimum of 7X coverage may be adequate if a reference genome is available, whereas a depth of 12X may suffice if genotyping de-novo. We base these conclusions on levels of SNP concordance (>95%) between gDNA and MDA as a function of coverage depth.
Finally, as alluded to above, MDA generally works best when using high molecular weight template. However, noninvasive sampling regimes are frequently confronted with the problem of highly degraded DNA (Taberlet et al. 1999, 2012b). Thus, an interesting and useful avenue for future research will be to ascertain if the potentially increased power to detect rare species by performing MDA on eDNA outweighs the bias introduced by the preferential amplification of long fragments. One solution to this trade-off would be to use WGA methods that aim to provide unbiased amplification of highly degraded template. For example, Wang et al. (2004) introduce a method termed Restriction and Circularization-Aided Rolling Circle Amplification that appears robust to moderate levels of degradation. In brief, the method first uses restriction enzymes to fragment the DNA between damage sites. Fragments are then circularized using DNA ligase and noncircularized fragments are discarded. The remaining double-stranded circularized DNA is denatured and amplified using φ29 polymerase, similar to traditional MDA (Dean et al. 2002). Early results show promise when working with highly degraded template, indicating that the method should be explored further.
Conclusions
The primary objective of this study was to establish a baseline estimate of bias levels combining MDA with ddRADseq under optimal conditions (>100 ng input DNA for MDA). Our results suggest that the efficacy of MDA scales with NGS applications including ddRAD-based studies. These conclusions are based on high concordance in both genome coverage and SNP genotypes when compared to raw gDNA samples. Future studies are needed to ascertain if similar results are obtained under suboptimal sampling conditions (e.g. low-quantity [<10 ng] and/or highly degraded template). We hope that MDA (and WGA methods more generally) will be found useful across numerous ecological and evolutionary applications. As many species and populations continue to be threatened by extinction, thus limiting field options for collecting biological samples, methods allowing genomic investigations from small noninvasive samples will be vital to implement adequate conservation measures.
Acknowledgements
We thank Erin Ehmke from the Duke Lemur Center for providing tissue samples. We thank P. Larsen for his insightful discussions regarding methods of statistical analysis and data visualization. J. Catchen provided invaluable advice regarding the use of Stacks. This research was funded by Duke University start-up funds to A.D. Yoder. This is Duke Lemur Center publication # 1283.
References
CB designed the experiment and performed all laboratory work. CB and CRC analysed the data. All authors contributed to the writing of the manuscript.
Data Accessibility
(i) Demultiplexed reads: NCBI Sequence Read Archive (Table S1, Supporting information). (ii) Stacks output files: DRYAD doi: 10.5061/dryad.83dc2.




