Deep learning to predict lymph node status on pre-operative staging CT in patients with colon cancer
S Bedrikovetski: BHSc (Hons); J Zhang PhD; W Seow MBBS; L Traeger MBBS, MTrauma; JW Moore MD, FRACS; J Verjans MD, PhD; G Carneiro MSc, PhD; T Sammour MBChB, FRACS, PhD.
Abstract
Introduction
Lymph node (LN) metastases are an important determinant of survival in patients with colon cancer, but remain difficult to accurately diagnose on preoperative imaging. This study aimed to develop and evaluate a deep learning model to predict LN status on preoperative staging CT.
Methods
In this ambispective diagnostic study, a deep learning model using a ResNet-50 framework was developed to predict LN status based on preoperative staging CT. Patients with a preoperative staging abdominopelvic CT who underwent surgical resection for colon cancer were enrolled. Data were retrospectively collected from February 2007 to October 2019 and randomly separated into training, validation, and testing cohort 1. To prospectively test the deep learning model, data for testing cohort 2 was collected from October 2019 to July 2021. Diagnostic performance measures were assessed by the AUROC.
Results
A total of 1,201 patients (median [range] age, 72 [28–98 years]; 653 [54.4%] male) fulfilled the eligibility criteria and were included in the training (n = 401), validation (n = 100), testing cohort 1 (n = 500) and testing cohort 2 (n = 200). The deep learning model achieved an AUROC of 0.619 (95% CI 0.507–0.731) in the validation cohort. In testing cohort 1 and testing cohort 2, the AUROC was 0.542 (95% CI 0.489–0.595) and 0.486 (95% CI 0.403–0.568), respectively.
Conclusion
A deep learning model based on a ResNet-50 framework does not predict LN status on preoperative staging CT in patients with colon cancer.
Introduction
Colon cancer is the fifth most diagnosed cancer amongst men and women worldwide. In 2020, over one million newly diagnosed cases and 576,858 deaths were attributed to this disease.1 The standard curative treatment remains complete resection of the primary tumour with regional lymph nodes (LN) and adjuvant chemotherapy in higher-risk patients.2 The presence of LN metastasis is a vitally important determinant of prognosis and treatment options.3, 4 Currently, these LNs are examined by specialist pathologists, with decisions about adjuvant therapy only possible after resection in patients without distant metastatic disease.5 In clinical practice, knowledge of preoperative LN involvement is rarely used given that neoadjuvant chemotherapy is typically only administered in patients with stage IV disease. Recently, the Foxtrot trial revealed that neoadjuvant chemotherapy can be delivered safely with the potential for pathological downstaging.6 However, this study included patients with a wide range of colon cancer staging. The limited diagnostic accuracy of pre-operative LN staging currently precludes the possibility of stratifying patients for neoadjuvant treatment.
Computed tomography (CT) is the most common imaging modality used in the preoperative staging of colon cancer. Despite excellent performance for the assessment of distant metastasis, the accuracy of preoperative assessment of LNs remains low, ranging from 54% to 64% using current diagnostic criteria based on size (LNs >1 cm).7-9 Several studies have attempted to apply different diagnostic criteria based on size, signal intensity, and morphology. However, the results of these studies are varied, and to date, there are no validated imaging criteria for the preoperative assessment of metastatic LNs.10-13
Artificial intelligence has demonstrated excellent diagnostic performance on preoperative LN staging in a variety of abdominopelvic malignancies.14 Deep learning as a subset of artificial intelligence is emerging as a more effective way to extract information from medical images in comparison with traditional models. Deep learning has the advantage of automatically and adaptively learning spatial hierarchies of features through its convolutional neural layers.15 However, evidence regarding the use of deep learning for predicting LN staging in patients with colon cancer is scarce. Therefore, this study aimed to develop and evaluate a deep learning model to predict LN status on preoperative staging CT in patients with colon cancer.
Materials and methods
Study design
This ambispective diagnostic cohort study is reported using the Artificial Intelligence in Medical Imaging (CLAIM) guidelines.16 The study protocol was approved by the Central Adelaide Local Health Network Human Research Ethics Committee (HREC/19/CALHN/73) and St Andrew's Hospital Human Research Ethics Committee (#116). This study was conducted in accordance with the principles of the Declaration of Helsinki. Informed consent was waived for all study participants.The goal of this study was to develop and evaluate a deep learning model used to predict LN status based on preoperative staging abdominopelvic CT in patients with colon cancer.
Data
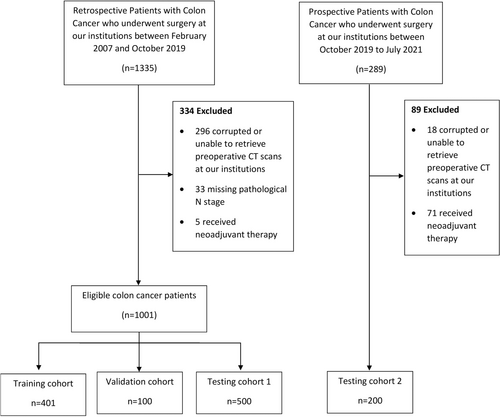
Patients diagnosed with colonic adenocarcinoma who underwent surgical resection with regional lymphadenectomy treated/referred to the Royal Adelaide Hospital or St. Andrews Hospital, South Australia, were eligible for inclusion. All included patients underwent standard unenhanced or contrast-enhanced CT preoperatively. As a result, some of the preoperative staging CT scans originated from the referring hospitals. The training cohort, validation cohort, and testing cohort 1 comprised 401, 100, and 500 retrospectively included patients treated between February 2007 and October 2019, respectively. Testing cohort 2 comprised 200 prospectively included patients treated between October 2019 and July 2021 with the same enrollment criteria. Exclusion criteria consisted of patients whose original CT scans were corrupted or not available, received neoadjuvant chemotherapy, or had missing pathological N stage. Baseline clinicopathological data including age, sex, tumour location, procedure type, and pathological TNM stage were extracted from a prospectively collected colorectal cancer database.
Ground truth
The ground truth for the N stage was determined by pathology assessment of the surgical specimen. Staging was based on the 8th edition of the American Joint Committee on Cancer (AJCC) TNM staging criteria.17
CT image acquisition and processing
All patients underwent 0.5–7 mm slice, standard unenhanced or post-intravenous contrast-enhanced preoperative CT of the abdomen and pelvis, with oral contrast or water as a negative contrast. We primarily analysed the portal venous phase CT images because of the clarity by which the LNs can be seen; however, we also analysed the few selected cases where only an arterial contrast-enhanced CT was available. A standard unenhanced CT was performed for patients with renal impairment or allergic to the intravenous contrast. Preoperative CT scans were exported from the Picture Archiving and Communication System (Carestream), or, through InteleViewer™ (Intelerad Medical Systems Inc, Montreal, Canada ) for CT scans performed in private imaging centers. Details regarding the CT systems are presented in Table S1. Axial plane sequences were isolated from the remainder of the CT images and anonymised using MicroDicom viewer (version 3.2.7; www.microdicom.com). Each axial plane CT sequence was assigned a binary label based on the ground truth (pN0 vs pN1-2).
Manual segmentation of regional LNs on axial slices was conducted by a science postgraduate student (S.B.) and a junior medical officer (W.S) trained and supervised by a senior colorectal surgeon (T.S.) who ensured the correct segmentation of the regional LNs during surgery using the ITK-SNAP software (version 3.6.0; www.itksnap.org) (Fig. S1).18 Regional LNs were segmented according to the anatomical location of the primary tumour. For right-sided tumours, segmentation included the mesenteric LNs around the ileocolic vessels (blood supply to the cecum and proximal ascending colon), right colic vessels (blood supply to the mid-distal ascending colon), and middle colic vessels (blood supply to the proximal to the mid-transverse colon) arising from the superior mesenteric vessels. For left-sided tumours, mesenteric LNs were segmented around the left colic vessels (blood supply to the distal third of the transverse colon, the splenic flexure, and descending colon) and sigmoid vessels (blood supply to the sigmoid colon) arising from the inferior mesenteric vessels. Manual LN segmentation was performed in the training and validation cohorts (n = 501) (Table S2).
Deep learning model
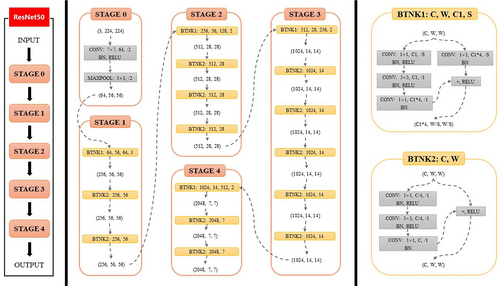
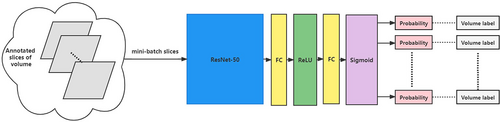
We proposed a convolutional neural network consisting of a segmentation ResNet-50 model and a classification ResNet-50 model to predict LN metastasis based on CT imaging.19 The ResNet-50 model consisted of 48 convolution layers, 1 MaxPool, and 1 Average Pool layer. In the segmentation task, the ResNet-50 (Fig. 1) was used as the encoder of the segmentation model, and the several transposed convolutions were followed by the residual blocks in the decoder (Fig. 2). We used the bilinear interpolation in the last layer to restore the feature map to the original resolution. The segmentation model played an assistant role in classification. We used the segmentation model to predict the positive slices in each volume which would be inputted into the classification model for diagnosis. The backbone (encoder) of the segmentation model was used to initialize the classification model. The ResNet-50 model, which has the same architecture as the segmentation encoder, was used in the classification task. The backbone was initialized by using the segmentation pre-trained weights. The pre-trained segmentation model was utilized for each volume to segment LNs and select 40 slices as the candidates for diagnosis. These candidates shared the same semantic label as the volume. The classification model took these slices as input to make the final decision. We used the binary cross-entropy loss to optimize the classification model (Fig. 3).

Performance evaluation
The prediction model was assessed by measuring the area under the receiver operating characteristic (AUROC) curve, accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV).
Statistical analysis
The parametricity of continuous measures was determined using the Shapiro–Wilk test. Normally distributed data were expressed as mean (standard deviation) and nonparametric data as median (range). Categorical measures were presented as frequencies and percentiles. A comparison of groups was performed using Pearson's chi-squared concerning categorical data. Exact or Monte Carlo methods were used for calculations depending on the table type and data count. One-way ANOVA or Kruskal–Wallis test was performed with respect to continuous data. A P value less than 0.05 was considered statistically significant. Statistical analysis was performed using IBM SPSS Statistics for Macintosh, version 28 (IBM Corp., Armonk, NY, USA) and MedCalc for Windows, version 20.027 (MedCalc Software, Ostend, Belgium).
Results
Baseline characteristics
A total of 1,201 patients (median (range) age, 72 (28–98) years; 653 (54.4%) male) were included in the study (Fig. 4). The clinicopathological characteristics for the training cohort (n = 401), validation cohort (n = 100), testing cohort 1 (n = 500) and testing cohort 2 (n = 200) are listed in Table 1. A significant difference was found between gender, tumour location, operation, pathological T stage and N stage, the total number of LN harvested, and the total number of positive LNs. A significant difference was also found in the types of scanners and the thickness of CT scan slices (Tables S1,S2).
| Variables | Training cohort (n = 401) | Validation cohort (n = 100) | Testing cohort 1 (n = 500) | Testing cohort 2 (n = 200) | P-value |
|---|---|---|---|---|---|
| Age, median (range), years | 74 (28–97) | 75 (30–91) | 71 (29–98) | 72 (29–94) | 0.26 |
| Gender | |||||
| Male | 241 (60.1) | 54 (54.0) | 251 (50.2) | 107 (53.5) | 0.03 |
| Female | 160 (39.9) | 46 (46.0) | 249 (49.8) | 93 (46.5) | |
| Tumour location | |||||
| Right | 222 (55.4) | 83 (83.0) | 302 (60.4) | 118 (59.0) | <0.001 |
| Left | 179 (44.6) | 17 (17.0) | 198 (39.6) | 82 (41.0) | |
| Operation | |||||
| Right hemicolectomy | 186 (46.4) | 43 (43.0) | 237 (47.4) | 91 (45.5) | <0.001 |
| Extended right/transverse colectomy | 32 (8.0) | 29 (29.0) | 56 (11.2) | 22 (11.0) | |
| Left hemicolectomy | 12 (3.0) | 5 (5.0) | 12 (2.4) | 3 (1.5) | |
| HAR | 130 (32.4) | 8 (8.0) | 148 (29.6) | 48 (24.0) | |
| Hartmann's | 9 (2.2) | 3 (3.0) | 12 (2.4) | 15 (7.5) | |
| Subtotal or total colectomy | 26 (6.5) | 10 (10.0) | 34 (6.8) | 16 (8.0) | |
| Proctocolectomy | 5 (1.2) | 0 (0.0) | 1 (0.2) | 2 (1.0) | |
| Other† | 1 (0.2) | 2 (2.0) | 0 (0.0) | 3 (1.5) | |
| pT stage | |||||
| T0/Tis | 8 (2.0) | 2 (2.0) | 17 (3.4) | 4 (2.0) | 0.003 |
| T1 | 49 (12.2) | 8 (8.0) | 59 (11.8) | 26 (13.0) | |
| T2 | 36 (9.0) | 5 (5.0) | 83 (16.6) | 20 (10.0) | |
| T3 | 220 (54.9) | 55 (55.0) | 244 (48.8) | 94 (47.0) | |
| T4 | 88 (21.9) | 30 (30.0) | 97 (19.4) | 56 (28.0) | |
| pN stage | |||||
| N0 | 261 (65.1) | 47 (47.0) | 324 (64.8) | 117 (58.5) | 0.004 |
| N1/2 | 140 (34.9) | 53 (53.0) | 176 (35.2) | 83 (41.5) | |
| Total no. of LNs harvested, median (range) | 16 (1–154) | 18 (1–60) | 18 (1–51) | 20 (1–124) | <0.001 |
| No. of positive LNs, median (range) | 0 (0–18) | 1 (0–32) | 0 (0–18) | 0 (0–12) | 0.001 |
- Data are presented as number (percentage) of patients unless otherwise indicated.
- HAR, high anterior resection; LNs, lymph nodes.
- † Other: ileocolic resections and total pelvic exenterations.
Performance of the deep learning model
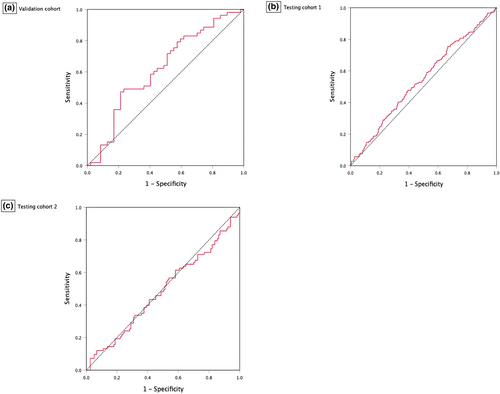
In the validation cohort, the deep learning model achieved an AUROC of 0.619 (95% CI 0.507–0.731) (Fig. 5). The deep learning model achieved a 96.2% (95% CI 87.0–99.5) sensitivity, 12.8% (95% CI 48.3–25.7) specificity, 57.0% (95% CI 46.7–66.9) accuracy, 55.4% (95% CI 52.4–58.4) PPV and 75.0% (38.9–93.4) NPV. For testing, the deep learning model yielded AUROC values of 0.542 (95% CI 0.489–0.595) in testing cohort 1 and 0.486 (95% CI 0.403–0.568) in testing cohort 2. The deep learning model showed high sensitivities of 96.6% (95% CI 92.7–98.7) and 91.6% (95% CI 83.4–96.5), low specificities of 5.2% (95% CI 3.1–8.3) and 6.0% (95% CI 2.4–11.9) and low accuracies of 37.4% (95% CI 33.1–41.8) and 41.5% (95% CI 34.6–48.7) in the testing cohort 1 and testing cohort 2, respectively. Of note, the model had PPVs of 35.6% (95% CI 34.8–36.5) and 40.9% (95% CI 39.0–42.8) and NPVs of 73.9% (95% CI 53.2–87.6) in the 2 testing cohorts, respectively (Table 2).
| Cohort | AUROC (95% CI) | Accuracy (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) |
|---|---|---|---|---|---|---|
| Validation cohort | 0.619 (0.507–0.731) | 57.0 (46.7–66.9) | 96.2 (87.0–99.5) | 12.8 (48.3–25.7) | 55.4 (52.4–58.4) | 75.0 (38.9–93.4) |
| Testing cohort 1 | 0.542 (0.489–0.595) | 37.4 (33.1–41.8) | 96.6 (92.7–98.7) | 5.2 (3.1–8.3) | 35.6 (34.8–36.5) | 73.9 (53.2–87.6) |
| Testing cohort 2 | 0.486 (0.403–0.568) | 41.5 (34.6–48.7) | 91.6 (83.4–96.5) | 6.0 (2.4–11.9) | 40.9 (39.0–42.8) | 50.0 (26.7–73.3) |
- AUROC, area under the receiver operating characteristic curve; NPV, negative predictive value; PPV, positive predictive value.
Discussion
In this ambispective diagnostic study, we attempted to develop a deep learning model to predict LN status on preoperative staging CT in patients with colon cancer. Our deep learning model showed a low predictive ability and reproducibility across validation and two different testing cohorts. Moreover, while the model had high sensitivity, it had very low specificity for malignant LNs. To our knowledge, this is the largest diagnostic study to use deep learning for the prediction of LN staging on preoperative CT imaging for patients with colon cancer.
Recently, two meta-analyses have shown that most artificial intelligence models used to predict LN staging in colorectal cancer are radiomics-based signatures.14, 20 However, this approach relies on predefined handcrafted features that carry inherent observer bias which may cause relevant information contained in the image to be missed or removed.21 Consequently, we developed a deep learning model to try to overcome this problem by automatically learning from LN segmentations and CT images, a model that might uncover subtle relations between LNs characteristics and metastatic potential.22, 23
Somewhat surprisingly, our preliminary deep learning model predicted the LN stage in patients with colon cancer with a higher AUROC (0.860 vs 0.486) than the present study.24 The discrepancy in diagnostic performance can be attributed to weak points of the preliminary study which included a smaller sample size (123 vs 1201 in the present study) and differences in deep learning architecture (DenseNet25 vs ResNet-50 in the present study). Compared with a recent radiomics study, the model in this study achieved a worse diagnostic performance with lower AUROC (0.486 vs 0.825), specificity (6% vs 86%), and accuracy (42% vs 79%).26 Importantly, the present study included only patients with colon cancer, however, our meta-analysis reported a higher diagnostic performance by radiomics models in comparison with the present study (AUROC 0.727 vs 0.486). Moreover, the significant difference in patient characteristics across cohorts may have affected the model's performance; however, reports suggest that datasets with diverse patient cohorts mitigate the bias of AI models.27, 28 Taken together, these results suggest that a radiomics-based approach using CT images is potentially more effective in predicting LN staging when compared to deep learning.
In clinical practice, the advantages of using a deep learning model over routine preoperative radiological LN staging include saving the substantial cost of radiology reporting and potentially improved accuracy leading to better targeting of treatment options. In comparison with previous studies, the sensitivity of our deep learning model may be higher but the model achieved consistently lower AUCs, accuracy, and specificities.11, 12, 29 This finding suggests that radiologists have a higher diagnostic capability in staging regional LNs on CT imaging in patients with colon cancer compared to the deep learning model in this study.
Several limitations of this study should be noted. First, segmentation of LNs was done manually, variability in segmenting LNs might lead to inconsistency in the extracted imaging features and subsequently influence the classification of LNs. In the future, this could be addressed with the use of automated segmentation tools which are rather less time-consuming and remove interobserver variability. Second, deep learning methods can be unpredictable when used with datasets with varying CT contrast and slice thickness from the training cohort, though different imaging parameters would enhance generalizability. Third, CT images were collected from different scanners, resulting in wide heterogeneity in imaging hardware and acquisition protocols. Regardless, selecting a single imaging protocol is an unrealistic reflection of daily clinical practice and would have made the results non-generalizable. Finally, the results of this study were from two institutions, so multicentre validation is required to assess reproducibility.
In conclusion, this study suggests a deep learning ResNet-50 model is not reliable in comparison with the current clinical standard in predicting LN status on preoperative staging CT in patients with colon cancer.
Acknowledgements
SA Health eHealth Innovation Grants Program (Project code 106637/66000), Colorectal Surgical Society of Australia and New Zealand (CSSANZ) Small Project Grant, CSSANZ Foundation Grant, and the University of Adelaide Divisional scholarship (a1649844).
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical approval
The study protocol was approved by the Central Adelaide Local Health Network Human Research Ethics Committee (HREC/19/CALHN/73) and St Andrew's Hospital Human Research Ethics Committee (#116).
Open Research
Data availability statement
The data that support the findings of this study are available on request from the corresponding author, SB. The data are not publicly available due to their containing information that could compromise the privacy of research participants.




