Temporal transcriptomic changes in willow leaves oviposited by Plagiodera versicolora
INTRODUCTION
Plants can establish a unique physiological state called the “Primed,” when exposed to a variety of environmental stresses. Under priming conditions, when challenged subsequently by pests, microbial pathogens, or abiotic stresses, plants typically respond faster or more strongly by activating defense responses (Mauch-Mani et al. 2017). These stress cues encompass various stimuli, such as insect feeding, sex pheromones, leaf volatile emissions from damaged adjoining plants, and insect egg deposition (Hilker & Fatouros 2015; Pashalidou et al. 2020).
The ability of plants to perceive and respond to insect oviposition has been confirmed in many species (Hilker & Fatouros 2015). Plants have developed a range of countermeasures in response to egg deposition. For example, Physalis leaves will grow neoplasms at the site of Heliothis subflexa egg deposition (Petzold-Maxwell et al. 2011). Molecular analyses revealed that insect oviposition can alter plant gene expression. The transcriptional changes induced by Pieris brassicae oviposition using Arabidopsis whole-genome DNA microarrays revealed that the expression of several genes associated with defense mechanisms increased (Little et al. 2007). Micromelalopha sieversi (Staudinger) egg deposition on poplar induces the expression of PR, NPR1/NIM1, and RPW8, which are associated with biological stress response (Guo et al. 2020).
Plagiodera versicolora Laicharting (Coleoptera, Chrysomelidae) is a globally distributed pest primarily affecting forest ecosystems (Ma et al. 2021; Xiong 2022). Recent studies have shown that P. versicolora egg deposition on Salix matsudana cv. ‘Zhuliu’ did not observe leaves' direct defense. However, it could alter the expression levels of transcription factors involved in metabolic responses and environmental stress. Three thousand seven hundred and ninety-five genes exhibited significant differential expression after 3 days of egg deposition by P. versicolora (Liu et al. 2023). However, the temporal transcriptomic changes in gene expression following oviposition by P. versicolora have not been reported.
Therefore, the present study was conducted on P. versicolora and willow. RNA-Seq was used to analyze the temporal dynamic changes of willow leaves after oviposition of P. versicolora, and the key candidate genes related to gene expression changes were explored. We aim to elucidate the interactions between insects and plants mediated by insect eggs and provide new insights into pest control.
MATERIALS AND METHODS
Six-month-old S. matsudana cv. ‘Zhuliu’ was purchased from Meng Yun Garden Shop (Wuhan, China) and kept in a greenhouse chamber at about 26 ± 1°C, 16-h light: 8-h dark photoperiod (16 L: 8 D), and 60% ± 5% relative humidity (RH). The P. versicolora adult individuals were collected from the surrounding Hankou Waterfront (Wuhan, China). They were placed in 25 cm × 15 cm × 10 cm plastic containers and provided with fresh willow leaves as their food source. The feeding conditions were the same as above (26 ± 1°C, 16 L: 8 D, 60% ± 5% RH).
In this experiment, we followed the method described by Liu et al. (2023) and used mated female P. versicolora to lay eggs on willow. The leaves were collected at 1, 6, 24, 48, and 72 h after treatment. Each treatment was replicated three times. The samples were immediately frozen in liquid nitrogen for the transcriptome analysis.
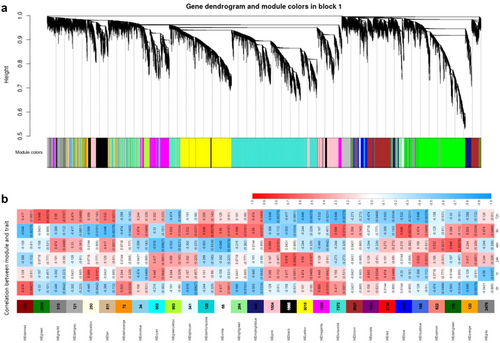
We proceeded with RNA extraction, cDNA library, Illumina sequencing, de novo assembly, and gene annotation using the method described by Liu et al. (2023). Weighted gene co-expression network analysis (WGCNA) was performed using the R package to generate an unsigned co-expression network based on topology overlap measurement for all sample data. This approach allowed us to identify and characterize the relationships between genes and their collective expression patterns. The soft threshold β = 16 was determined when the fitting curve was close to 0.9 for the first time. Furthermore, hub genes are defined as genes that are highly interconnected within a key module. For each of these hub genes, we individually visualized their network relationships, providing a comprehensive view of their interactions within the network. The networks were visualized using Cytoscape v.3.10.0 (NCBI SRA accession: PRJNA1007945). To explore the biological relevance of the key module, comprehensive functional enrichment analyses were performed using gene ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG). Results with corrected P-value < 0.05 were considered significantly enriched.
RESULTS AND DISCUSSION
The de novo transcriptome of the egg deposition willow comprised 27 757 459, 28 305 283, 27 789 461, 28 266 426, 27 471 488, and 28 584 960 clean reads at 0, 1, 6, 24, 48, and 72 h, respectively, with a total of 266 763 transcripts obtained and assembled into 132 483 unigenes.
To investigate the temporal transcriptional changes of willow leaves to oviposition by P. versicolora, using WGCNA for analysis, by constructing a dendrogram, we were able to observe distinct branches representing 28 unique module eigengenes (Fig. 1a). There was only one co-expression module, MElightyellow, which comprised genes exhibiting high expression levels at 1 h (Fig. 1b). We consider that these genes are involved in the swiftness of the willow transcriptome response to insect egg deposition. The MEorange and MEroyalblue modules exhibit similar expression patterns after oviposition treatment. They show positive high expression at 6, 24, and 48 h and negative expression at 1 and 72 h, which is associated with the adaptation to the process of egg deposition (Fig. 1b). The MEgreen module displayed significant up-regulation exclusively at 72 h, potentially acting as a signal for larvae to initiate hatching (Fig. 1b).
The significant GO terms enriched by eigengenes of module lightyellow included “Metabolic and cellular process” and “Biological regulation and response to stimulus” (Fig. 2a). KEGG pathways with the highest unigene representations were “Plant hormone signal transduction” and “Phenylpropanoid biosynthesis” (Fig. 2c). Meanwhile, the eigengenes in module green were mainly enriched in “Metabolic and cellular process” and “Biological regulation and response to stimulus” (Fig. 2b). KEGG pathways with the highest unigene representations were “Carbon fixation in photosynthetic organisms,” followed by “Circadian rhythm” and “Glyoxylate and dicarboxylate metabolism” (Fig. 2d). These findings imply that egg deposition of P. versicolora induces changes in transcript expression related to metabolic responses and plant stress. The process of “Metabolic and cellular process” increases the amount of plant secondary metabolites, impairing larvae performance and reducing the risk of plant feeding by insects. The process of “Biological regulation and response to stimulus” can promote plant growth or early flowering to complete the life cycle as early as possible, and this stress-avoidance response in plants has very important biological implications in insect–plant interactions.
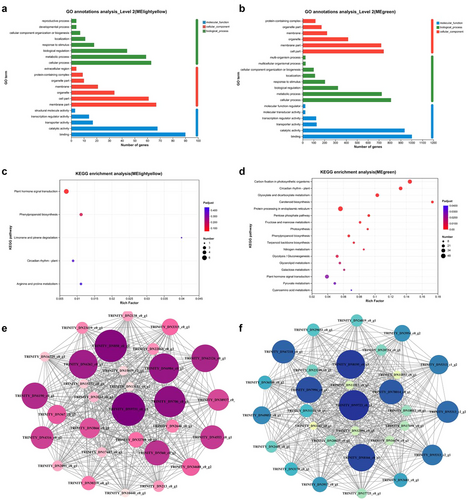
To identify hub genes, we focused on the top 30 genes with the highest intra-modular connectivity. In the lightyellow module, yellow stripe-like protein, pathogenesis-related genes transcriptional activator, and auxin transporter protein are highly expressed (Fig. 2e), and in the green module, including b-box zinc finger protein, detoxification protein, and late elongated hypocotyl (Fig. 2f). These key genes may be involved in various biological processes such as regulation of transcription, immune response, and regulation of circadian rhythms. These results indicate that willow leaves sense changes in the timing of egg deposition by expressing different types of genes.
In conclusion, this study was particularly focused on the temporal transcriptomic changes in willow leaf oviposition by P. versicolora. We aimed to investigate the molecular responses of willow using RNA-Seq and WGCNA methods and accurately localize the plant defense genes and functional enrichment pathways associated with insect egg deposition. This can also provide a new perspective for understanding the interaction between insects and plants.