The first chromosome-level genome assembly and transcriptome sequencing provide insights into cantharidin production of the blister beetles
Abstract
Blister beetles (Coleoptera: Meloidae) produce a natural defensive toxin cantharidin (CTD), which has been used for various cancer treatments and other diseases. Currently, the lack of chromosome-level reference genomes in Meloidae limits further understanding of the mechanism of CTD biosynthesis and environmental adaptation. In this study, the chromosome-level genome assembly of Mylabris phalerata was generated based on PacBio and Hi-C sequencing. This reference genome was about 136.68 Mb in size with contig N50 of 9.17 Mb and composed of 12 chromosomes. In comparison to six other Coleoptera insects, M. phalerata exhibited multiple expanded gene families enriched in juvenile hormone (JH) biosynthetic process pathway, farnesol dehydrogenase activity, and cytochrome P450, which may be related to CTD biosynthesis. Consistently, the transcriptomic analysis suggested the “terpenoid backbone biosynthesis” pathway and “the juvenile hormone” as putative core pathways of CTD biosynthesis and presented eight up-regulated differential expression genes in male adults as candidate genes. It is possible that the restricted feeding niche and lifestyle of M. phalerata were the cause of the gene family's contraction of odorant binding proteins. The ABC transporters (ABCs) related to exporting bound toxins out of the cell and the resistance to the self-secreted toxins (e.g. CTD) were also contracted, possibly due to other self-protection strategies in M. phalerata. A foundation of understanding CTD biosynthesis and environmental adaptation of blister beetles will be established by our reference genome and discoveries.
INTRODUCTION
Meloidae species, commonly known as blister beetles (Huang et al. 2016), produce a sesquiterpenoid defensive toxin, cantharidin (C10H12O4, CTD). CTD is an important traditional Chinese medicine for treating various diseases like malign sores, dropsy, and warts (Wang 1989). Previous studies have identified that CTD has anticancer properties (Li et al. 2017), which could be used as an alternative to anticancer drugs. Moreover, CTD is widely used in plant protection owing to its fungicidal and herbicidal properties (Carrel & Eisner 1974; Matsuzawa et al. 1987).
Over the past few decades, studies of blister beetles, a natural source of CTD, mainly focused on the genetic mechanism of CTD biosynthesis. The genomes of Hycleus cichorii and H. phaleratus have been assembled to provide genomic resources for investigating CTD biosynthesis (Wu et al. 2018). Furthermore, genome-wide analyses of Mylabris aulica indicated possible genes and pathways associated with CTD biosynthesis (Guan et al. 2019). Gene family evolution analysis based on Epicauta chinensis genome identified two candidate genes (CYP4TT1 and phytanoyl-CoA dioxygenase) that possibly contributed to CTD biosynthesis (Tian et al. 2021). Additionally, different blister beetle species may produce CTD through different genes or functional pathways. Therefore, due to the complexity of CTD biosynthesis, its genetic mechanism requires in-depth investigations based on higher-quality blister beetle genomes.
The quality of genome assembly has an impact on the reliability of bioinformatic analyses in areas such as comparative genomics, functional genomics, and population genomics (Mishra et al. 2022). Previous studies on blister beetle genomes used sequencing methods like Illumina, nanopore, or PacBio (Tian et al. 2021). Although these assemblies have been used as reference genomes for population genomics and comparative genomics, their fragmented and incomplete nature makes it challenging to study chromosome evolution, a key driver of speciation and species evolution. While some genes related to CTD biosynthesis have been discovered, there is a possibility of identifying new genes for a better understanding of the entire mechanism with higher-quality genomes. The Hi-C method, which identifies chromosomal interactions based on chromosome conformation capture, has been used to group and organize contigs based on their physical proximity in the genome (Belton et al. 2012). In this study, we have generated the first chromosome-level genome of the Meloidae species, Mylabris phalerata, which is of significant importance in Chinese traditional medicine. We utilized PacBio sequencing and Hi-C technology to achieve this, and we aimed to conduct comparative genomics analyses using this chromosome-scale genome to identify the key genetic factors involved in CTD biosynthesis in blister beetles. The genetic data and discoveries obtained through our study will offer fresh perspectives into the genetic indicators of CTD biosynthesis.
CTD is mostly synthesized by adult male beetles, while adult females only obtain CTD through mating (Huang et al. 2016). Investigations into the mechanism of sexual dimorphism in CTD biosynthesis have been conducted through a variety of physiological and biochemical studies. Comparative transcriptomics is a very effective and efficient approach to exploring this regulation mechanism. An examination of Mylabris cichorii’s transcriptome and expression profiling identified the potential role of the mevalonate (MVA) pathway and juvenile hormone (JH) biosynthesis in CTD biosynthesis (Huang et al. 2016). Analyses of Epicauta tibialis transcriptomes, comparing them, revealed seven genes (differential expression genes [DEGs]) that had been increased in expression in male adults. These genes, which were located in the terpenoid biosynthesis pathway, could be the cause of the sexual dimorphism in CTD production (Du et al. 2021). However, comparative analysis of transcriptome data using high-quality reference genomes is rather limited. The gene expressions were examined based on the chromosome-level genome generated in this study to determine key genes and pathways associated with sexual dimorphism in CTD biosynthesis.
MATERIALS AND METHODS
Genome sequencing and assembly
In 2021, adult specimens of M. phalerata were collected from Puge County, Sichuan Province, China. The PacBio Sequel II platform in Novogene Co., Ltd. (Beijing, China) was used to sequence the library that had passed the qualification. The resulting HIFI reads were then used to assemble the primary genome using HiFiasm with default settings (Cheng et al. 2021). BUSCO (Benchmarking Universal Single-Copy Orthologs) was used to analyze the genome's completeness (Simão et al. 2015).
Hi-C scaffolding
The preparation of the Hi-C library followed a previously published method for insects (Shi et al. 2019). Initially, whole tissues from each individual were fixed using formaldehyde. A restriction enzyme was employed to cross-link and digest the chromatin, and in situ labeling was conducted, wherein a biotinylated residue was appended to the 5' overhangs of the DNA fragments, followed by end repair. After ligation, the DNA fragments containing biotin were extracted and sheared. Illumina NovaSeq 6000 platform was then used to sequence the libraries, using a 150-bp paired-end layout. After filtering, the remaining unique sequences were utilized for Hi-C assembly and the generation of visual interaction maps using 3D-DNA (Dudchenko et al. 2017). To obtain the final assembly results with Juicebox, manual adjustments were made to correct any misordered or misoriented sequences.
Genome annotation
Uncovering simple sequence repeats (SSRs) in the M. phalerata genome, the software Krait (with default parameters) was utilized (Du et al. 2018). These SSRs can serve as useful loci for population genetic studies. RepeatModeler and repeatmasker were employed for repeat annotation (Zhou et al. 2022). For predicting protein-coding genes (PCGs) in M. phalerata, a comprehensive strategy was adopted and integrated using the EVidenceModeler pipeline (Haas et al. 2008). GlimmerHMM, AUGUSTUS, and geneMark were employed in de novo gene prediction. Homologous gene annotation was performed using GeneWise, with protein sequences from four species: Tribolium castaneum, Dendroctonus ponderosae, Anoplophora glabripennis, and Drosophila melanogaster (Zhou et al. 2022). Employment of Program to Assemble Spliced Alignments (PASA) enabled the acquisition of gene structures from transcriptome data (Haas et al. 2003). BUSCO was used to assess the PCGs’ completeness, and functional annotation was conducted in line with our previous study (Zhou et al. 2022)
Phylogenetic tree construction and positive selection
OrthoFinder was utilized for gene family identification (Zhou et al. 2022) for 11 species (Table S1, Supporting Information). Initially, one-to-one orthologous genes were aligned using mafft (Katoh & Standley 2013) with default parameters. After the employment of PAL2NAL (Suyama et al. 2006), ambiguously aligned blocks were removed using Gblocks (Castresana 2000), resulting in the concatenation of the remaining CDS alignments for each gene family into a supergene per species. RAxML (Stamatakis 2014) and MCMCtree (Yang 2007) were used for phylogenetic and divergence analyses, respectively.
Gene family expansion and contraction
This analysis was conducted using CAFÉ (De Bie et al. 2006; Zhou et al. 2023) and was based on the constructed phylogenetic tree. A random birth and death process model, along with a P-value threshold of 0.05, was employed to identify significantly expanded or contracted gene families within each lineage represented on the phylogenetic tree. KOBAS was utilized to enrich the above genes (Zhou et al. 2022).
Manual annotation of specific genes
For comparisons of gene families, we followed the methodology from our previous study (Zhou et al. 2019). These sequences were then processed through TBLASTN (Kent 2002), SOLAR (Ashburner et al. 2000), and GeneWise (Zhou et al. 2022) according to our previous methodology (Zhou et al. 2019). With hmmscan (http://hmmer.org), we sought out genes in the translated protein sets obtained. With IQ-TREE (Trifinopoulos et al. 2016), a maximum likelihood (ML) algorithm, with 1000 iterations of bootstrapping and the LG+R8 best-fit model, was utilized to construct the unrooted phylogenetic tree.
RNA sequencing and identification of differentially expressed genes
Following Du et al. (2021), we conducted RNA sequencing of the whole body without the gut. Data filtering and assessment, genome mapping, and differential expression analysis were performed. Since the number of reads in RNA-seq data can be influenced by technical errors, sequencing depth, and gene length, we applied the TMM algorithm in edgeR (Robinson & Oshlack 2010) to normalize the gene counts. We then employed the standardized gene expression matrix to analyze the gene expression disparities between male and female M. phalerata. DEGs were enriched via the R package clusterProfiler (Yu et al. 2012).
RESULTS AND DISCUSSION
Genome sequencing and assembly
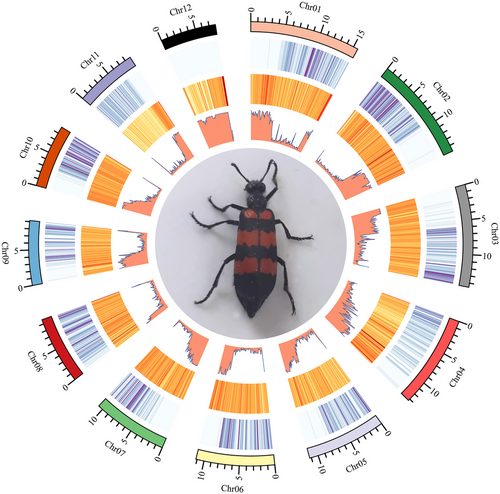
A total of 41.41 Gb of HiFi data were obtained for the de novo genome assembly of M. phalerata. We obtained 136.68 Mb M. phalerata genome in size (contig N50: 9.17 Mb; scaffold N50: 11.24 Mb). These scaffolds accounted for 99.53% of the whole genome assembly orientated and anchored to 12 chromosomes (Fig. 1). The genome size of M. phalerata in this study was larger than H. cichorii, H. phaleratus, and M. aulica (Wu et al. 2018; Guan et al. 2019). The genome completeness of M. phalerata was estimated to be 92.1% complete BUSCOs covered by the genome, of which 91.7% were single copies and 0.4% were duplicated.
Genome annotation
The obtained genome assembly of M. phalerata contained 56.81 Mb (41.57% of the genome) repetitive elements (Table 1). M. phalerata’s repetition elements in the genome were larger than E. chinensis genome and two Hycleus genomes (Wu et al. 2018; Tian et al. 2021), while lower than M. aulica (Guan et al. 2019). The variation among species was estimated to be related to genome sizes (Chalopin et al. 2015). DNA elements (22.63 Mb, 16.56%) were the most abundant categories of transposable elements, while short interspersed nuclear elements (SINEs, 64.30 b, 0.13%) were the least. The high proportion of unclassified elements (24.04 Mb, 17.59%) may be due to the lack of studies on the repeats of Coleoptera. Table S2, Supporting Information, provides a comprehensive overview of the perfect SSR's total and length, which were 29 540 and 678 865 base pairs, respectively.
| Type of repeats | Subfamily | Number of elements | Length occupied (bp) | Percentage in the genome (%) |
|---|---|---|---|---|
| SINEs | 285 | 64 294 | 0.05 | |
| ALUs | 0 | 0 | 0.00 | |
| MIRs | 1 | 106 | 0.00 | |
| LINEs | 5848 | 2 577 147 | 1.89 | |
| LINE1 | 871 | 430 094 | 0.31 | |
| LINE2 | 637 | 63 681 | 0.05 | |
| L3/CR1 | 214 | 62 582 | 0.05 | |
| LTR elements | 6710 | 7 499 027 | 5.49 | |
| ERVL | 4 | 254 | 0.00 | |
| ERVL-MaLRs | 0 | 0 | 0.00 | |
| ERV_classI | 113 | 8072 | 0.01 | |
| ERV_classII | 81 | 5178 | 0.00 | |
| DNA elements | 34 741 | 22 632 235 | 16.56 | |
| hAT-Charlie | 716 | 283 167 | 0.21 | |
| TcMar-Tigger | 104 | 26 777 | 0.02 | |
| Unclassified | 55 859 | 24 039 373 | 17.59 |
- DNA, DNA transposons; LINE, long interspersed nuclear elements; SINE, short interspersed nuclear elements; LTR, long terminal repeated elements.
The total of 9787 protein-coding genes with 89.5% completeness was obtained by combining de novo, homolog-based, and transcript-based sequences, 9639 (98.49%) of which were functionally annotated using five public protein databases (Table S3, Supporting Information). Of these annotated genes, 23 genes were in the “insect hormone biosynthesis” pathway (map00981) (Fig. S1, Supporting Information). These two pathways were considered to be responsible for CTD synthesis in blister beetles (Huang et al. 2016). The CTD biosynthesis-related genes allow the species to replace or fill the current medical market for blister beetles.
Gene family and phylogenetic analysis
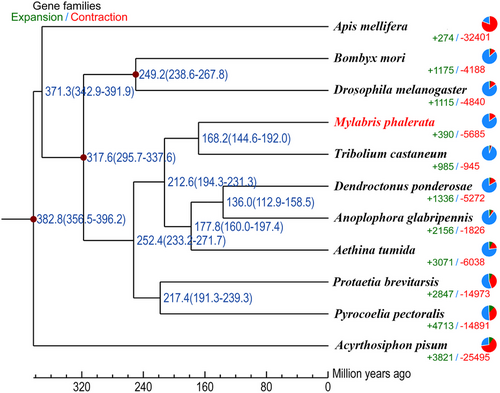
Comparative genomics analysis was undertaken among 11 insect species. Identification of gene families laid the foundation for comparative genomics analysis, which facilitated a better understanding of the evolutionary traits among different species. We identified 40 554 gene families, of which 6017 gene families were commonly shared in the branch of five Coleoptera species and 435 gene families were specific to M. phalerata. A phylogenetic tree was constructed based on 514 one-to-one orthologous genes. M. phalerata was sister to T. castaneum and grouped with other Coleoptera species. Divergence time analysis revealed that M. phalerata diverged from the recent common ancestor approximately 168.2 Mya, similar to that between Meloidae species and T. castaneum while more ancient than the previous estimation (around 104 Mya) (Guan et al. 2019) (Fig. 2).
Gene family expansion
Comparative genomic investigation was performed to infer the evolutionary history of M. phalerata. We identified a total of 390 expanded and 5685 contracted gene families in M. phalerata. Gene family expansion is considered an important mechanism for phenotypic diversity and evolutionary adaption to the environment (Harris & Hofmann 2015). GO enrichment of expanded genes indicated JH biosynthetic process pathway (GO:0006718) and farnesol dehydrogenase activity (GO:0047886), which were possibly related to CTD biosynthesis. KEGG enrichment uncovered the insect hormone biosynthesis pathway and terpenoid backbone biosynthesis, both of which were essential for CTD biosynthesis (Tian et al. 2021). M. phalerata might promote CTD production through certain gene family expansions for the adaptation to hostile environments.
RNA sequencing and putative genes and pathways related to the sexual dimorphism in the cantharidin biosynthesis
Comparative transcriptional analyses were performed between male and female M. phalerata for their differences in CTD production. A total of 69 Gb of paired-end data were created for M. phalerata, and the percentage of Q30 was about 87.3–92%. Correlation analysis showed that different biological repetitions had a high consistency and were clustered together (Fig. 3a). A principal component analysis (PCA) plot showed that 28.6% and 23.0% of the total gene expression variance from these 10 samples were explained by the first two principal components (Fig. 3b). The PC1 clearly separated these samples into two groups corresponding to males and females, indicating the differential expression patterns resulting from sex.
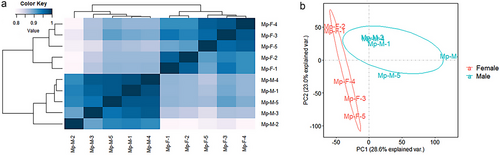
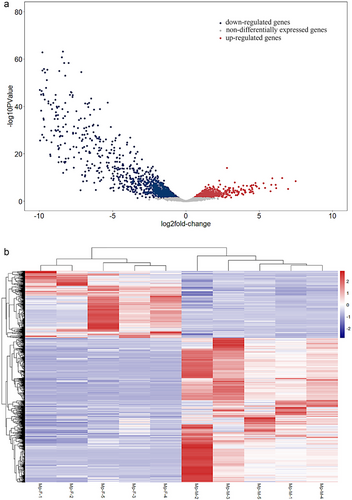
Figure 4a reveals 1481 genes that expressed differently (471 up-regulated genes and 1010 down-regulated genes) and were visualized in a heatmap. The up-regulated DEGs in males showed a significant correlation (P value < 0.05) with “terpenoid metabolic process,” “Cytochrome P450,” “ABC transporters,” “regulation of defense response to fungus,” “regulation of Toll signaling pathway,” and “positive regulation of antimicrobial peptide production.” The up-regulated genes involved in the “terpenoid metabolic process” and P450s revealed their functional correlation with CTD synthesis. ABC transporters may be important for sequestering potential CTD precursors to avoid auto-intoxication (Fratini et al. 2021). According to sex-specific resource investment strategies, female insects had a more sensitive immune system than males when infected by pathogens. However, males may have a higher baseline level for immune maintenance (Barthel et al. 2015). Functional analysis indicated that female adults may have stronger detoxification ability, which was similar to the finding in Bombus huntii (Xu et al. 2013). However, there remains a limited understanding of the CTD synthesis process in insects. Some genes associated with MVA and MEP/DOXP were estimated to be related to CTD biosynthesis in M. aulica (Guan et al. 2019), whereas most studies on the blister beetles argued the MEP pathway out of the candidate pathway related to CTD synthesis (Du et al. 2021) and our results confirmed this finding.
MVA pathway could control the de novo synthesis of isoprenoid cantharidin verified by isotope-labeling research (Huang et al. 2016). Most critical enzyme genes of the MVA pathway were found and annotated in M. phalerata, such as acetyl-CoA C-acetyltransferase, hydroxyl-methylglutaryl-CoA synthase, mevalonate kinase, and phosphomevalonate kinase. Additionally, atoB, HMGCS, and HMGCR showed high expression in males. Similar high expressions were observed in many other blister beetles. These genes were involved in catalyzing acetyl-CoA into mevalonate, and silencing of HMGR can significantly reduce CTD production in male E. chinensis (Lu et al. 2016). Together, this evidence indicates their correlation with CTD biosynthesis.
Farnesol is the precursor of CTD, thus we searched genes in the farnesol-related branch. To start, we pinpointed farnesyl diphosphate synthase and isopentenyl-diphosphate delta-isomerase with significantly higher levels in males. These two are key enzymes of farnesol metabolism and geranyl diphosphate formation. Previous observations on the transcript expression of FDPS and IDI also showed higher levels in males. Therefore, FDPS and IDI represented plausible candidate genes for the synthesis of CTD in blister beetles. ICMT, STE24, FACE2, and FNTB were identified from the trans-farnesol branch. These genes were relatively conserved in blister beetles. Among them, STE24 showed significantly up-regulated expression, indicating its correlation with the sexual dimorphism and the biosynthesis of CTD. Consistent with that, the expression of this gene was found to be twofold higher in male beetles (20–25 day-old) than in females (Huang et al. 2016).
Three enzyme genes upstream of the “Juvenile hormone” pathway, which control JH synthesis, were identified by us, such as farnesyl diphosphate phosphatase, NADP+-dependent farnesol dehydrogenase, and aldehyde dehydrogenase (NAD+). FOHSDR and ALDH were the members of expanded gene families, implying that they worked as the underlying driving force of the evolution of CTD synthesis. The up-regulated expression of ALDH in the JH pathway was also found in E. tibialis (Du et al. 2021). JH III acid diol was likewise considered a precursor of CTD (Jiang et al. 2019). Here, we only detected the JH esterase, not including JH epoxide hydrolase (JHEH) reported in a previous study (Fratini et al. 2021). JHE's essentiality in controlling JH degradation and its consequences on the precursor of CTD cannot be overstated. Therefore, JHE may partly mediate CTD biosynthesis.
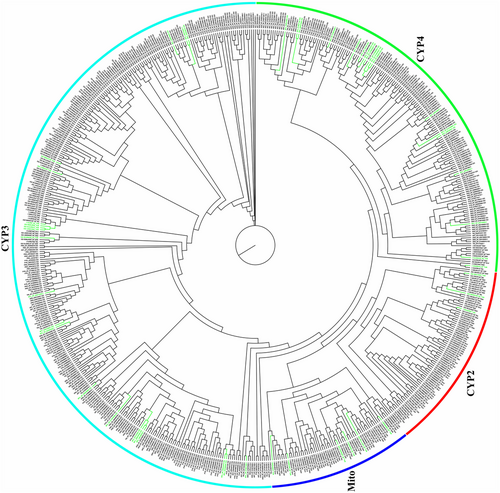
Cytochrome P450 gene family
We found 48 putative members in M. phalerata, which was the least number among these six beetles. According to the phylogenetic tree, we identified 3 CYP2, 21 CYP3s, 18 CYP4s, and 6 mitochondrial genes (Mito) in the genome of M. phalerata (Fig. 5). Mito and CYP2 clade were basic components of P450s, despite small proportions. The conservation of Mito and CYP2 across families was found to be highly significant, and they were mainly associated with the biosynthesis or metabolism of endogenous compounds, particularly JHs and ecdysteroids (Shi et al. 2022). Although M. phalerata lost quite a few members of CYP3 and CYP4 that were found in other Coleoptera insects and had single members in most lineages, there remained slight booms within some small lineages that had a close relationship with T. castaneum. A wide range of evidence has linked multiple members of CYP4s to CTD biosynthesis (the cytochrome P450 gene CYP4BM1, CYP4TT1, and the homologs) (Jiang et al. 2019; Tian et al. 2021). CYP4BM1 exhibited high expression in the midgut or reproductive tract of blister beetles (Lydus trimaculatus and Mylabris variabilis) and showed a positive correlation with CTD production. Its homologous gene CYP4TT1 was identified in Epicauta chinensis, and its expression displayed a similar correlation with CTD contents (Tian et al. 2021).
Chemoreception and detoxification systems
In this study, there were 154 chemosensory genes identified in M. phalerata (Fig. 6). Compared to other Coleoptera beetles, the number of odorant binding proteins (OBPs) underwent obvious contraction in M. phalerata, even less than the stenophagous D. ponderosae. OBPs have key functions in the recognition and binding of odor molecules and delivering them to odorant receptors on dendritic membranes (Andersson et al. 2019). Larvae of M. phalerata were reported to feed on eggs of the grasshopper Chondracris rosea rosea De Geer (Orthoptera: Acridiidae) in China (Zhu et al. 2006). OBPs vary in size between insect taxa due to different rates of gene gain and loss, and this variation is estimated to be linked to specific lifestyles and adaptations to the environment (Andersson et al. 2019). The loss of OBPs may be related to their special lifestyle and restricted feeding niche, suggesting a potential adaptive strategy in M. phalerata.

There were 48 P50s, 38 ABCs, 31 CCEs, 15 GSTs, and 22 UGTs annotated in M. phalerata (Fig. 6), among which CYPs, ABCs, CCEs, and GSTs showed significant contraction compared to other Coleoptera beetles. The ABCs took up major roles in the detoxification system of M. phalerata. Phase III enzyme ABC transporters or other membrane transport proteins were linked with exporting bound toxins out of the cell, and the latest evidence suggests their resistance to self-secreted toxins (e.g. CTD) (Fratini et al. 2021). It was reported that blister beetles developed self-protection strategies by toxin sequestration mechanisms to prevent auto-intoxication due to the release and accumulation of CTD in the body.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant number: 31960117) and the Miaozi Project in Science and Technology Innovation Program of Sichuan Province (grant number: 2022006).
CONFLICT OF INTEREST
The authors declare no competing interests.




