Contributions of Lake Erie and Lake St. Clair Walleye Populations to the Saginaw Bay, Lake Huron, Recreational Fishery: Evidence from Genetic Stock Identification
Abstract
Genetic stock identification analyses were conducted to determine spawning population contributions to the recreational fishery for Walleyes Sander vitreus in Saginaw Bay, Lake Huron. Two spawning population groups were considered: (1) the Tittabawassee River, which has been identified as the largest source of spawning Walleyes for Saginaw Bay; and (2) an aggregate of six spawning populations from Lake Erie and Lake St. Clair that were found to be genetically similar. Overall, the Lake Erie and Lake St. Clair spawning populations were estimated to comprise approximately 26% of the Walleye recreational harvest in Saginaw Bay during 2008–2009. Contribution levels were similar for the 2 years in which genetic samples were collected. Contributions from the Lake Erie and Lake St. Clair spawning populations to the harvest of age-5 and older Walleyes were greater during summer (31.8%; SE = 6.2%) than during late winter and spring (6.0%; SE = 3.7%). Conversely, contributions from the Lake Erie and Lake St. Clair spawning populations to the harvest of age-3 and age-4 fish were fairly similar between seasons (late winter and spring: 31.2%, SE = 6.7%; summer: 41.7%, SE = 5.6%), suggesting that younger Walleyes migrate earlier or reside in Saginaw Bay for extended periods. Our finding that one-quarter of the Saginaw Bay recreational harvest of Walleyes comprises fish from Lake Erie and Lake St. Clair has important management implications, as policies for one lake may have bearing on the other lake—one of the challenges associated with managing migratory fish species. Fisheries management in the Laurentian Great Lakes has a history of being highly coordinated and cooperative among the states and province bordering the individual lakes. Results from this study suggest that cooperation may need to be expanded to account for fish movement between lakes.
Received October 28, 2014; accepted February 11, 2015
Species that move long distances relative to the scale at which policy decisions are made present a number of challenges for fisheries management. First, when populations mix during periods of harvest, there is an increased risk of overexploitation if the populations vary in productivity (Frank and Brickman 2000; Kell et al. 2009; Ying et al. 2011). Additionally, movement can affect the performance of methodologies for estimating important dynamic rate functions of populations (Quinn et al. 1990; Goethel et al. 2011; Li et al. 2015). The fisheries management process is further complicated when fish traverse jurisdictional boundaries, as fisheries in one area can depend on populations and systems over which managers have no jurisdictional authority (Hilborn and Sibert 1998). Although issues related to migratory fish populations have been highlighted in the management of marine fishes, freshwater fisheries management also confronts challenges stemming from migratory behavior in fishes, particularly for large and interconnected systems (Molton et al. 2012, 2013; Vandergoot and Brenden 2014; Li et al. 2015).
The Walleye Sander vitreus is one example of a freshwater fish species that is known to exhibit migratory behavior (Smith et al. 1952; Rasmussen et al. 2002; Wang et al. 2007). Migration is typically seasonal and often relates to spawning. Walleyes spawn in the spring, and multiple studies have documented strong spawning site fidelity (Forney 1963; Olson et al. 1978; Hayden et al. 2014). Prior to spawning, mature Walleyes stage on or near the spawning grounds (Munger 2002). After spawning, fish make directed movements toward summer feeding grounds to replenish their energy resources (Wang et al. 2007). The distance between summer feeding grounds and spawning locales varies widely depending on the spawning area and the stock, but it is during this postspawning period that Walleyes may move considerable distances (Rasmussen et al. 2002; Wang et al. 2007; Hayden et al. 2014), possibly as a result of temperature preferences or prey availability (Kershner et al. 1999; McParland et al. 1999; Zhao et al. 2011). Hayden et al. (2014) reported that some Walleyes moved more than 350 km in a 30-d period after spawning. As water temperatures decline in the fall, fish begin returning to areas near spawning grounds in preparation for the upcoming spawning season.
Lake Erie and Saginaw Bay in Lake Huron support two of the largest Walleye fisheries in the world (Schneider and Leach 1979; Baldwin et al. 2009). Since 2000, annual Walleye harvest in Lake Erie has averaged close to 3 million fish, with harvest split fairly equally between recreational (44%) and commercial (56%) fisheries (WTG 2014). Prior to the fishery collapse in the mid-1940s, the annual yield of Walleyes in Saginaw Bay averaged approximately 458,000 kg (Baldwin et al. 2009). Improvements in water quality and initiation of a Walleye stocking program led to the emergence of a recreational fishery in Saginaw Bay during the 1970s (Fielder 2002). Between 1986 and 2005, the annual recreational harvest of Walleyes in Saginaw Bay (including some tributary fisheries that exploited fish from the bay) averaged approximately 81,000 fish (Fielder et al. 2014). Beginning in the early 2000s, alterations in prey fish abundances led to a change in the Lake Huron fish community (Riley et al. 2008; He et al. 2015). Walleye reproductive success in Lake Huron increased substantially beginning in 2003 as a consequence of declining Alewife Alosa pseudoharengus abundances (Fielder et al. 2007), and the increased reproductive success ultimately led to the cessation of Walleye stocking (Fielder and Thomas 2014). Since 2006, annual recreational harvest of Walleyes has averaged approximately 227,000 fish (Fielder et al. 2014; T. Kolb, Michigan Department of Natural Resources [MDNR], unpublished data). In 2009, the Walleye population in Saginaw Bay met recovery targets and ultimately was declared to have recovered (Fielder and Thomas 2014). Because of the recovery, there is new emphasis on the management of Walleyes throughout Lake Huron, and the increased abundance has elevated their importance as part of the lakewide predator suite affecting predator–prey balance (He et al. 2015).
Lake Erie and Lake Huron are connected via the Detroit River, Lake St. Clair, and the St. Clair River, which are collectively referred to as the “Huron–Erie Corridor” (Figure 1). Studies that have involved the tagging of spawning Walleyes in Lake Erie during the spring have documented their migration into the Huron–Erie Corridor and Lake Huron (Ferguson and Derksen 1971; Todd and Haas 1993; Wang et al. 2007). Ferguson and Derksen (1971) documented such movements for fish as young as age 1. For some regions of Lake Erie, as much as 10% of the Walleyes (largely males) that are tagged when spawning have been found to migrate into the Huron–Erie Corridor and Lake Huron (Todd and Haas 1993; Wang et al. 2007; Vandergoot and Brenden 2014), although movement rates vary among years (Wang et al. 2007). Tagging studies have provided estimates of migration rates into Saginaw Bay, but determining the overall contribution of Lake Erie Walleyes to the Saginaw Bay recreational harvest has not been previously attempted. Even at low migration rates, the contribution from Lake Erie has the potential to be large given the abundance estimates of the two populations (Saginaw Bay: 1.4–4.0 million age-2 and older [age-2+] Walleyes from 1986 to 2011, Fielder and Bence 2014; west-central basin of Lake Erie: 20–90 million age-2+ Walleyes from 1980 to 2014, WTG 2014). In the mid-1990s, Walleye commercial fisheries in the Ontario jurisdictional waters of Lake Huron were estimated to be supported primarily (67–72%) by fish emigrating from Lake Erie (McParland et al. 1999), which demonstrates that Lake Erie can make major contributions to a Walleye fishery in Lake Huron. Contribution levels to Lake Huron and Saginaw Bay may have changed recently as a consequence of declining prey availability in Lake Huron. Knowledge of how many Lake Erie Walleyes are contributing to the Saginaw Bay recreational harvest would be beneficial given the fishery management implications of such a contribution.
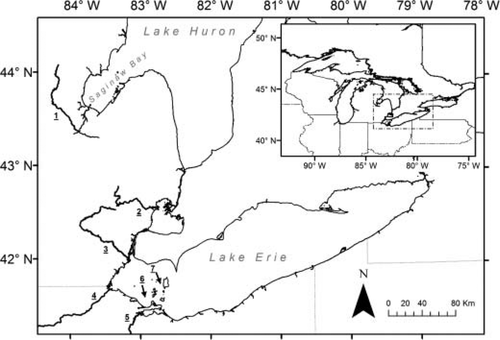
Map of Saginaw Bay (Lake Huron) and Lake Erie, showing locations of individual Walleye spawning populations from which genetic samples were collected in relation to the location of the Saginaw Bay mixture fishery (1 = Tittabawassee River, Lake Huron; 2 = Clinton River, Lake St. Clair; 3 = Huron River; 4 = Maumee River; 5 = Sandusky River; 6 = Toussaint Reef; 7 = Chicken Island Reef). Inset shows the locations of Saginaw Bay, Lake Huron, and Lake Erie relative to the other Laurentian Great Lakes.
The purpose of this study was to (1) estimate the contribution of Walleyes from Lake Erie and Lake St. Clair spawning groups to the Saginaw Bay, Lake Huron, recreational fishery; and (2) evaluate how contribution to the recreational harvest varied seasonally and by age. To accomplish this, we generated genetic profiles for several Walleye spawning populations from Lakes Huron, St. Clair, and Erie and for samples of fish harvested from Saginaw Bay in February–August of 2008 and 2009; we then used genetic stock identification analyses and estimated fish ages to estimate the contribution levels for the assessed spawning populations.
METHODS
Sampling methods.
Fin clips were obtained from individuals representing seven spawning populations: Tittabawassee River (Saginaw Bay, Lake Huron; n = 96), Clinton River (Lake St. Clair; n = 48), Chicken Island Reef (Lake Erie; n = 48), Huron River (Lake Erie; n = 50), Maumee River (Lake Erie; n = 48), Sandusky River (Lake Erie; n = 48), and Toussaint Reef (Lake Erie; n = 48; Figure 1). Tissue samples from the Clinton River were collected as part of a study conducted in the late 1990s (K. T. Scribner, unpublished data); samples from the other spawning populations were collected in spring 2010 as part of routine monitoring of Walleye spawning populations (i.e., sampling was conducted on spawning areas during the spawning season) by the MDNR, Ohio Department of Natural Resources, and Ontario Ministry of Natural Resources. Tissue samples were dried and stored in paper envelopes. The Tittabawassee River population was the only Lake Huron spawning population from which tissue samples were collected because it was the only population assessed during that year. Furthermore, the Tittabawassee River population has traditionally been identified as the single largest spawning population contributing to the Saginaw Bay Walleye fishery (Fielder 2014), and past research has found that genetic differentiation between Lake Huron spawning populations is low relative to differences between Lake Erie sources (Stepien et al. 2010).
Walleye tissue samples from the Saginaw Bay recreational fishery were obtained from archived dorsal spines collected for aging in 2008 (n = 258) and 2009 (n = 279) during the annual creel program. We chose to process tissue samples from these years because in comparison with other years, the samples were more evenly distributed among months. All of the Walleyes for which we analyzed tissue samples had been aged by using dorsal spines. The dorsal spines were aged by one experienced reader, which was deemed sufficient due to the growth rates of Saginaw Bay Walleyes (Fielder and Thomas 2014). Ages of fish from which tissue samples were collected ranged from 3 to 15 years (age 3: 19% of sampled fish; age 4: 29%; age 5: 27%; age 6: 16%; age 7: 5%; age 8+: 4%). Samples from the creel program were available for the months of February–August (February: 13% of sampled fish; March: 16%; April: 10%; May: 11%; June: 16%; July: 20%; August: 14%). The sex of fish was only recorded for 42% of the available samples. Among those samples for which sex was identified, the percentage female was 34% in 2008 and 48% in 2009.
Laboratory analysis.
QIAGEN DNeasy kits (QIAGEN, Germantown, Maryland) were used to extract DNA from fin tissue samples according to the manufacturer's protocols. Quantification of DNA was accomplished by using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, Delaware). All DNA samples were diluted to a concentration of 20 ng/μL for use in PCR.
Individuals were genotyped at 10 microsatellite loci: Svi4, Svi17, Svi18, and Svi33 (Borer et al. 1999); SviL2, SviL5, SviL6, and SviL8 (Wirth et al. 1999); and Svi6 and Svi7 (Eldridge et al. 2002). Loci Svi18 and Svi33 were amplified together in a single reaction, as were Svi4 and Svi17. The PCRs were conducted in 10-μL volumes using 40 ng of DNA and included PCR buffer (10-mM tris-HCl at pH 8.3, 50-mM KCl, 0.01% gelatin, 0.01% NP-40, and 0.01% Triton-X 100); 0.2-mM deoxynucleotide triphosphates (dNTPs) for single-locus reactions or 0.24-mM dNTPs for reactions amplifying two loci; varying concentrations of MgCl2; and infrared, fluorescently labeled forward primers and unlabeled reverse primers. Locus-specific details of PCR amplification conditions are presented in Supplementary Table S.1 (available in the online version of this article). The PCR cycling conditions consisted of one cycle of an initial denaturation step for 2 min at 94°C; a varying number of cycles of 45 s at 94°C, 1 min at the annealing temperature, and 1 min at 72°C; and one cycle of 1 min at the annealing temperature followed by 10 min at 72°C. Deviations from these cycling conditions included (1) an extension time of 1 min 15 s for the Svi4–Svi17 locus pair during the main amplification cycles; and (2) a 1-min denaturation step during the main amplification cycles as well as a final cycle consisting only of incubation for 5 min at 72°C for Svi7.
The PCR products were separated by size on a denaturing 6.5% polyacrylamide gel and were visualized using the LI-COR 4300 DNA Analyzer (LI-COR Biosciences, Lincoln, Nebraska). All genotypes were independently scored by two experienced laboratory personnel, and 10% of the samples were randomly selected and re-genotyped at all 10 loci. Genotype scores were compared with the original scores to derive an empirical estimate of scoring error, which was 0.68%.
Data analysis.
Even though Walleyes from the spawning populations were collected during the spawning season, there still was the potential for collected individuals to be strays (i.e., first-generation migrants) from other populations. We identified potential strays by using GeneClass2 (Cornuet et al. 1999). Strays were identified via the method recommended by Paetkau et al. (2004). This entailed calculating the ratio of two likelihoods for each individual: (1) the likelihood that an individual of a particular genotype originated from the spawning area where it was collected; and (2) the maximum such likelihood considered over all possible spawning areas (Paetkau et al. 1995). A Monte Carlo resampling method was then used to generate a null distribution of straying for each spawning population against which observed likelihood ratios could be compared (Paetkau et al. 2004). Null distributions were generated by simulating 10,000 individuals from each population. A threshold P-value of 0.01 was used to identify strays.
For each spawning population, the number of alleles, allelic richness, observed heterozygosity (Ho), and expected heterozygosity (He) were calculated for each locus. Number of alleles and allelic richness were also calculated across all spawning populations combined. Each locus for each spawning population was tested for conformity to Hardy–Weinberg equilibrium (HWE) using Genepop version 4.2 (Raymond and Rousset 1995; Rousset 2008). Genepop was also used to test for linkage equilibrium (i.e., random association of alleles) between each pair of loci for each spawning population. Significance values for HWE and linkage equilibrium tests were generated with Markov chain algorithms by resampling 1,000 iterations/batch for 100 batches. Bonferroni corrections were used to adjust the α levels as a result of simultaneous testing (HWE: α = 0.05/[10 loci × 7 populations] = 0.000714; linkage equilibrium: α = 0.05/[45 locus pairs × 7 populations] = 0.00016). Multiple genetic fixation indices (FST = mean genetic divergence between pairs of spawning populations; FIS = mean genetic differentiation within spawning populations; FIT = deviation in the total sample; Weir and Cockerham 1984) were calculated using FSTAT version 2.9.3.2 (Goudet 1995, 2001). Overall genetic differentiation among spawning populations was calculated using pairwise FST values (Weir and Cockerham 1984). Significance of pairwise FST values was assessed by randomization (total number of permutations = 1,000).
The ability of our spawning population data set to accurately assign individual Walleyes to their location of collection was evaluated by estimating the stock composition of simulated single-population samples (i.e., 100% mixture simulations). Simulations were conducted in ONCOR (Kalinowski et al. 2007), which implements the simulation approach of Anderson et al. (2008), and involved repeated (number of iterations = 1,000) generation of mixtures that comprised only fish from one of the spawning populations. Bootstrapped mixture sample sizes were set equal to 200 fish, and spawning population sample sizes were set equal to the actual sample sizes for the populations. Our target accuracy level for the 100% mixture simulations was 90% for each spawning population (Seeb and Crane 1999; Beacham et al. 2012). If the target of 90% accuracy was not achieved, then spawning populations with the lowest pairwise FST values were grouped together and the 100% mixture simulations were conducted anew. This process was repeated until the 90% accuracy target was achieved in all spawning population groups.
To verify that the number of spawning population groupings that were identified from the 100% mixture simulations was appropriate and that our baseline data did not exclude other genetically differentiated spawning populations contributing to the Saginaw Bay fishery, we pooled individuals from the spawning populations and the mixture, and we used STRUCTURE version 2.3.1 (Pritchard et al. 2000; Falush et al. 2003) to conduct an admixture analysis that probabilistically assigned individuals to putative genetic clusters. Admixture analyses assumed uncorrelated allele frequencies and involved 100,000 Markov chain–Monte Carlo iterations with a 50,000-iteration burn-in period. Analyses were conducted with an assumed number of populations or clusters (K) ranging from 1 to 10 and using 10 replicates for each value of K. To determine the number of clusters that best fit our pooled data, we used STRUCTURE HARVESTER (Earl and vonHoldt 2012) to summarize the STRUCTURE output estimates of the likelihood of each K given the data for each replicate.
As noted below in the Results, only two spawning population groups were ultimately identified from the 100% mixture and STRUCTURE analyses. As an additional means for evaluating the accuracy and precision of genetic stock identification analyses conducted using these two spawning populations as baselines, we used ONCOR to simulate mixture fisheries that consisted of varying proportions of these populations, and we compared the estimated contributions with the actual contributions. Population contributions were varied from 10% to 90% in 10% increments, and simulations were conducted in a similar manner to the 100% mixture simulations (i.e., 1,000 iterations over observed spawning population sample sizes; and a mixture sample size of 200 fish). We refer to these simulations as “realistic mixture” simulations.
Examination of results from the 100% mixture and realistic mixture simulations suggested that there was a consistent relationship between the mean contribution estimates for the spawning populations and the contribution levels assumed in the simulations. We used this relationship to bias-correct the contribution estimates from actual genetic stock identification analyses. Bias correction involved fitting a quadratic polynomial model to the simulation results (independent variable = mean contribution estimate for a spawning population from the simulation analyses; dependent variable = assumed contribution estimate for a spawning population in the simulations). We chose a quadratic relationship rather than a linear relationship because there appeared to be a nonlinear trend in the results. We chose to fit a model to the results from only one of the spawning populations, and the contribution estimates for the other population were then corrected by subtraction. Alternatively, we could have developed correction equations for each of the two spawning population groups, but this would have resulted in contribution estimates not summing to 100%, and there was practically no difference (<0.4%) in the estimates obtained from the two approaches.
Genetic stock identification of the tissue samples from the Saginaw Bay recreational fishery based on the spawning population groups identified from the 100% mixture simulations was performed in the Statistics Program for Analyzing Mixtures (SPAM) version 3.7 (ADFG 2003). We used SPAM rather than ONCOR for the mixture analyses because SPAM outputs the log-likelihood for the fitted model, which we could then use to test differences in spawning population contribution estimates (see below). The SPAM and ONCOR programs implement similar procedures for genetic stock identification, and the estimated contribution levels differed by less than 0.3% between the two software packages in all cases when applied to our data, so our use of SPAM over ONCOR had little bearing on our overall conclusions. The mixture fishery data were analyzed in several different arrangements to determine the consistency of spawning population contributions (1) for all Saginaw Bay Walleyes (all samples grouped together); (2) by sampling year (2008 versus 2009); (3) by age-group (ages 3 and 4 versus age 5+); (4) by season (February–May samples versus June–August samples); and (5) by age-group and season combined (levels as described in numbers 3 and 4 above). We tested for statistical significance of differences in spawning population contributions among the different factor levels by using likelihood ratio tests (Reynolds and Templin 2004). For the likelihood ratio tests, we used an asymptotic theory approach rather than a resampling-based approach because we only had two spawning population groups and because the contribution estimates were generally not close to parameter space boundaries (Reynolds and Templin 2004).
Our categorization of age-groups and seasons was influenced in part by sample size limitations but was also informed by the behavior of Great Lakes Walleyes. Based on tagging studies, migration rates of age-3 and age-4 Walleyes into Lake Huron were believed to be lower than those of age-5+ fish (Wolfert 1963; Ferguson and Derksen 1971; Wang et al. 2007; Fielder 2014). With regard to seasonal differences, the spawning of Walleyes in the Great Lakes generally commences in mid- to late-March and continues throughout much of April and into early May, with staging occurring prior to actual spawning. As a result, it was hypothesized that Walleyes would not migrate into Lake Huron until early June.
RESULTS
Seven Walleyes collected from the spawning populations were identified as being strays (i.e., first-generation migrants) from one of the other populations. Straying was identified as occurring between spawning populations from different lakes as well as between spawning populations within Lake Erie. Straying occurred from the Huron and Clinton rivers to the Tittabawassee River; from Chicken Island Reef to the Huron River; from the Huron River to the Maumee River; from the Maumee River to the Sandusky River; and from the Tittabawassee River to Toussaint Reef. All seven identified strays were excluded from analyses summarizing genetic characteristics and from the 100% mixture simulations that were used to assess the potential accuracy of mixture analyses. However, because of the spawning population groupings we considered for actual mixture analyses, individuals that were identified as straying between Lake St. Clair and Lake Erie or between populations within Lake Erie were retained for the realistic mixture simulations and for actual mixture analyses.
Combined across spawning populations, the number of alleles observed per locus ranged from 6 (Svi18) to 24 (Svi7; Table S.2). When calculated for individual spawning populations, the number of alleles per locus was fairly consistent among the populations (Table S.2), differing by no more than four alleles between populations. However, as evidenced by differences between the allele counts for all populations combined and the allele counts for individual populations, some alleles were unique to a subset of spawning populations (e.g., for SviL5, the number of alleles for all populations combined was 23, whereas the range in number of alleles for individual populations was 14–18). Values of Ho and He were generally in close agreement, although there were some locus × spawning population combinations for which relative differences in Ho and He were as much as 20% (e.g., Svi18 for Toussaint Reef; Table S.2). Each spawning population conformed to HWE expectations at each locus, with an error rate of 0.000714 after Bonferroni correction (Table S.2). Out of the 315 possible spawning population × locus pair combinations that were evaluated, three combinations were found to be in linkage disequilibrium: (1) Svi17 and SviL2 for the Tittabawassee River population; (2) Svi4 and Svi6 for the Sandusky River population; and (3) Svi7 and Svi6 for the Chicken Island Reef population. Because linkage disequilibrium was only identified in a single spawning population for each of these locus combinations, we did not feel it necessary to exclude any of the loci for which linkage disequilibrium was detected.
Combined across loci, genetic fixation indices were as follows: 0.015 (95% confidence interval [CI] = 0.008–0.024) for FST, 0.026 (95% CI = 0.005–0.048) for FIT, and 0.011 (95% CI = −0.006 to 0.030) for FIS (fixation indices for individual loci are presented in Table S.2). The most genetically distinctive spawning population was the Tittabawassee River population (mean pairwise FST value = 0.026; Table 1). The next most distinctive spawning populations were the Chicken Island Reef and Toussaint Reef populations (mean pairwise FST value for each population = 0.013; Table 1). Each of the remaining populations had mean pairwise FST values ranging from 0.007 to 0.008 (Table 1).
| Population | Tittabawassee River | Clinton River | Huron River | Maumee River | Sandusky River | Toussaint Reef |
|---|---|---|---|---|---|---|
| Clinton River | 0.0178* | |||||
| Huron River | 0.0172* | 0.0020* | ||||
| Maumee River | 0.0234* | 0.0046* | 0.0055* | |||
| Sandusky River | 0.0287* | 0.0027 | 0.0022 | −0.0012 | ||
| Toussaint Reef | 0.0399* | 0.0039 | 0.0089 | 0.0075 | 0.0024 | |
| Chicken Island Reef | 0.0286* | 0.0108* | 0.0103* | 0.0014 | 0.0114* | 0.0183* |
The 100% mixture simulations for the seven original spawning populations indicated that the expected accuracy of population-specific composition was high for the Tittabawassee River (≈89%). However, accuracy levels for the other spawning populations ranged from 21% to 50%. Although we attempted a number of different spawning population groupings, we only achieved our 90% target accuracy level through a grouping that consisted of just two spawning populations: (1) the Tittabawassee River population and (2) all Lake Erie and Lake St. Clair populations combined (hereafter, “Lake Erie–Lake St. Clair population group”). Under this grouping, accuracy from the 100% mixture simulations was 98.4% (95% CI = 95.1–100.0%) for the Lake Erie and Lake St. Clair spawning populations and 90.1% (95% CI = 83.9–95.5%) for the Tittabawassee River population. The Lake Erie–Lake St. Clair population group conformed to HWE expectations at each locus (Table S.2).
The unstructured admixture analysis (STRUCTURE) of the pooled mixture and spawning population data supported the findings from the 100% mixture simulations. Both the mean loge probability of the data given K clusters and the rate of change in that mean probability (Evanno et al. 2005) indicated that just two spawning population groupings gave rise to the baseline and mixture data. The rate of change (i.e., ΔK) was 190.9 for two spawning populations, whereas the second-largest rate of change was 12.2 for four populations.

where ŷ represents the estimate of the true contribution by the Lake Erie–Lake St. Clair population group; and x represents the mean estimated contribution from the simulation analyses (Figure 2). This equation yielded a near-perfect fit (r2 > 0.99) to the 100% mixture and realistic mixture simulation results. As a result, we used this equation to correct the estimated contributions of the Lake Erie–Lake St. Clair population group. As previously indicated, the corrected contribution estimates for the Tittabawassee River population were obtained by subtracting the Lake Erie–Lake St. Clair group's contribution from 100%. All contribution estimates reported hereafter are corrected values, although it should be noted that the results of the likelihood ratio tests are based on uncorrected estimates.
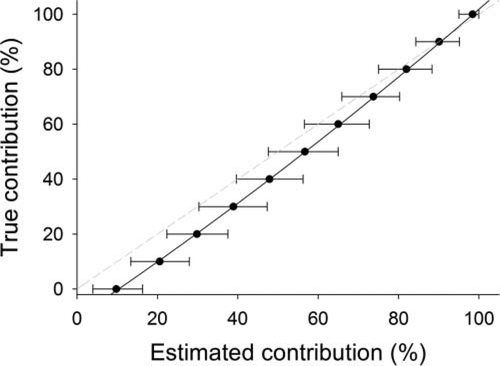
Results from 100% mixture and realistic mixture simulations conducted on Walleye genetic data from the Lake Erie and Lake St. Clair spawning populations and the Tittabawassee River (Lake Huron) spawning population; simulations were used to evaluate the accuracy and precision of genetic stock identification analysis. Plotted results are for the combined Lake Erie–Lake St. Clair spawning population group (error bars = 95% confidence limits from bootstrapping; solid black line = bias correction relationship estimated from the mixture simulations; dashed gray line = 1:1 reference line).
The proportional contribution estimates for the pooled sample from the Saginaw Bay recreational fishery were approximately 26% for the Lake Erie–Lake St. Clair population group and 74% for the Tittabawassee River spawning population (Table 2). The contributions in 2008 and 2009 were similar (Table 2) and did not significantly differ between the two years (χ2 = 1.93, df = 1, P = 0.165). When contributions to different age-groups in the Saginaw Bay harvest were examined, the Lake Erie–Lake St. Clair group's contribution was significantly greater for age-3 and age-4 Walleyes (38%) than for age-5+ fish (17%; χ2 = 8.54, df = 1, P = 0.003). When contributions to seasonal harvest were analyzed, the Lake Erie–Lake St. Clair group's contribution was significantly greater during June–August (37%) than during February–May (15%; χ2 = 9.70, df = 1, P = 0.002).
| Scenario | Level | Sample size | Lake Erie and Lake St. Clair | Tittabawassee River | SE |
|---|---|---|---|---|---|
| Pooled | 537 | 26.1 (35.3) | 73.9 (64.7) | 2.7 | |
| Year | 2008 | 258 | 28.9 (37.9) | 71.1 (62.1) | 4.1 |
| 2009 | 279 | 23.4 (32.8) | 76.6 (67.2) | 3.7 | |
| Age-Group | Ages 3 and 4 | 255 | 37.6 (45.8) | 62.4 (54.2) | 4.3 |
| Age 5 and older | 282 | 16.1 (26.0) | 83.9 (74.0) | 3.4 | |
| Season | February–May | 269 | 15.0 (24.9) | 85.0 (75.1) | 3.5 |
| June–August | 268 | 37.4 (45.7) | 62.6 (54.3) | 4.2 | |
| Age-Group × Season | Ages 3 and 4, February–May | 99 | 31.2 (40.1) | 68.8 (59.9) | 6.6 |
| Ages 3 and 4, June–August | 156 | 41.7 (49.5) | 58.3 (50.5) | 5.6 | |
| Age 5 and older, February–May | 170 | 6.0 (16.3) | 94.0 (83.8) | 3.7 | |
| Age 5 and older, June–August | 112 | 31.8 (40.5) | 68.2 (59.5) | 6.2 |
Overall, the differences between the age-group × season combinations were statistically significant (χ2 = 17.35, df = 4, P = 0.002). Contributions from the Lake Erie–Lake St. Clair population group to age-3 and age-4 fish in the Saginaw Bay harvest were similar between seasons (Table 2), with no significant difference (χ2 = 1.45, df = 1, P = 0.229). However, the Lake Erie–Lake St. Clair group's contributions to age-5+ harvested fish were significantly different between seasons (χ2 = 7.36, df = 1, P = 0.007): June–August contributions were approximately 26% greater than February–May contributions (Table 2). During February–May, contributions from the Lake Erie–Lake St. Clair population group to age-3 and age-4 fish in the harvest were significantly greater than contributions to age-5+ fish (χ2 = 6.36, df = 1, P = 0.012). However, contributions to the two age-groups were not significantly different for the June–August period (χ2 = 1.29, df = 1, P = 0.256).
DISCUSSION
Although multiple tagging studies have previously documented the migration of Walleyes from Lake Erie and Lake St. Clair into Lake Huron (Ferguson and Derksen 1971; Todd and Haas 1993; Wang et al. 2007), the magnitude of contributions to the Saginaw Bay recreational fishery has been difficult to quantify. Tagging studies conducted in Lake Erie and Lake St. Clair have been skewed toward males and particular ages, and only a few spawning populations have been tagged each year (Vandergoot and Brenden 2014). Furthermore, tag shedding rates during studies on Lake Erie have differed depending on the agency that conducted the tagging (Vandergoot et al. 2012); if not accounted for, such differences could affect contribution estimates based on the recovery of tags. As previously indicated, given the relatively large sizes of the Lake Erie and Lake St. Clair populations, they could contribute considerable numbers of fish even at low migration rates. Statistical catch-at-age models have been developed for Walleyes in both Lake Erie and Lake Huron, so estimates of abundance are available (Fielder and Bence 2014; WTG 2014); however, the data sets that are used to fit these models reflect the true migration of Walleyes between the lakes, and thus there is the potential for bias in abundance estimates depending on how migration is treated in the models.
Because of the difficulties outlined above, genetic studies such as this one provide important new information—not available from tagging studies alone—that can be used to assess the overall contribution of Lake Erie and Lake St. Clair Walleyes to the Lake Huron fishery as long as (1) there is sufficient genetic divergence to permit reliable differentiation among spawning populations and (2) reasonable efforts have gone into inventorying possible spawning populations. Given the low genetic divergence and the level of inaccuracy observed in the 100% mixture simulations, it is likely not possible to accurately determine the levels of contribution from each of the six original spawning populations from Lake Erie and Lake St. Clair. However, by combining the genetics data into a single Lake Erie–Lake St. Clair population group, our contribution estimates ostensibly account for any noninventoried spawning populations that are genetically similar to the inventoried populations. Strange and Stepien (2007) and Stepien et al. (2010) assessed genetic similarity for a total of 13 Lake Erie and Lake St. Clair Walleye spawning populations; they found that spawning populations in rivers along the southern shore of Lake Erie's western and central basins were linked by high connectivity and gene flow. Therefore, we anticipate that noninventoried spawning populations in this area of Lake Erie would be reflected in the Lake Erie–Lake St. Clair group's contribution estimates. Strange and Stepien (2007) and Stepien et al. (2010) did find that other Lake Erie spawning populations were genetically divergent from those along the southern shore of the western and central basins and that some of these populations were more closely related to Lake Huron spawning populations based on neighbor-joining tree analysis. Thus, if Walleyes from these other Lake Erie populations were included in the Saginaw Bay mixture fishery sample, it is possible that they were assigned to the Tittabawassee River population. This would mean that Lake Erie–Lake St. Clair group contribution estimates would be negatively biased relative to the actual contributions. Conversely, positive biases in Lake Erie–Lake St. Clair contribution estimates could result if the Saginaw Bay recreational fishery included contributions from Lake Huron spawning populations that were more genetically similar to Lake Erie and Lake St. Clair spawning populations than to the Tittabawassee River population. For example, reef spawning by Walleyes in Saginaw Bay was historically important (Schneider and Leach 1979), and efforts to restock Saginaw Bay in the early 1990s included some Walleyes reared from western Lake Erie reef-spawning populations (MDNR, unpublished data). Fielder (2002) collected reef-spawning Walleyes in Saginaw Bay, and those fish were subsequently found to be genetically dissimilar to Tittabawassee River spawners based on mitochondrial DNA genetic markers (Billington et al. 1998). More recent analyses (K. T. Scribner, unpublished data) showed that Walleyes collected from Saginaw Bay reefs and from the Tittabawassee River were genetically similar to Walleyes from the Muskegon River (a Lake Michigan tributary), a primary progenitor stock used in restoration. Thus, the Tittabawassee River and Saginaw Bay Walleyes were not genetically similar to other “native” Lake Huron fish or to Lake Erie–Lake St. Clair fish. We do not know (1) whether reef-spawning Walleyes in Saginaw Bay could be mistaken for Lake Erie–Lake St. Clair fish or (2) the size of such reef-spawning populations, but these factors could influence our results. Contributions from other Walleye spawning populations in Lake Huron are deemed unlikely, as most are considered to be in a depressed state and are affiliated with tributaries to the North Channel or Georgian Bay, and migration from these areas to Saginaw Bay is limited (Fielder et al. 2010; Fielder and Bence 2014). Furthermore, based on the work of Stepien et al. (2010), Lake Huron spawning populations that have been genetically profiled cluster together phylogenetically, so there is little obvious reason why Walleyes from Lake Huron populations would assign to the Lake Erie–Lake St. Clair population group.
The results from our research suggest that Walleyes from Lake Erie and Lake St. Clair comprised between 6% and 42% of the harvest (depending on age-group and season) in the Saginaw Bay recreational fishery during 2008 and 2009. Although age-5+ Walleyes from Lake Erie and Lake St. Clair primarily contributed to the Saginaw Bay recreational fishery during the summer months, age-3 and age-4 Walleyes from Lake Erie and Lake St. Clair contributed nearly as much during late winter and spring as during the summer. This suggests that age-3 and age-4 Walleyes either migrate earlier than older fish or that they reside in Saginaw Bay for an extended period of time prior to undertaking seasonal migrations. Overall, we found that Lake Erie–Lake St. Clair Walleyes comprised approximately one-quarter of the Saginaw Bay recreational fishery for this species. In 2008 and 2009, total annual Walleye harvest in Saginaw Bay was approximately 300,000 fish, which equates to roughly 80,000 harvested Walleyes originating from Lake Erie and Lake St. Clair. Additional studies will be needed to determine the degree of interannual variability in contribution levels. We believe that the use of archived tissue (e.g., scale) samples to determine the contribution levels occurring prior to the Alewife collapse would be beneficial for assessing how contributions have changed as the Lake Huron food webs have changed. Future genetic stock identification analyses of Walleyes in Lake Huron would clearly benefit from efforts to identify and genetically inventory the potentially contributing spawning populations, particularly those associated with midwater reefs in Saginaw Bay.
The estimated Lake Erie–Lake St. Clair population contributions to the Saginaw Bay recreational fishery are consistent with anticipated contributions based on estimates of recreational exploitation rates in Lake Huron, abundance levels in Lake Erie, and migration rates between the two lakes. Applying the recreational exploitation rates from Fielder and Bence (2014) to the estimated abundance of age-3+ Walleyes in Lake Erie's central and western basins (WTG 2014), a migration rate of between 1% and 2% would be needed to achieve a 26% contribution to the Saginaw Bay recreational fishery; this is within the range of previously estimated migration rates from Lake Erie (Vandergoot and Brenden 2014). Still, there are several factors that could have caused our contribution estimates to be either inflated or unrepresentative of Walleye population composition in Saginaw Bay. As discussed above, estimates could be inflated if there were Lake Huron spawning populations contributing to the Saginaw Bay recreational fishery that were more genetically similar to the Lake Erie and Lake St. Clair spawning populations than to the Tittabawassee River population (e.g., putative reef-spawning strains derived from earlier stocking of Lake Erie fish). Estimated contribution levels in the fishery harvest might also deviate from the composition of the at-large Saginaw Bay Walleye stock if there are factors that make Lake Erie–Lake St. Clair fish more vulnerable to harvest (e.g., greater catchability; or differences in age-specific vulnerability due to differences in growth pattern). Presently, we do not have evidence that these factors influenced our contribution estimates, but we nevertheless believe that it is important to identify potential biases in our estimates.
The impetus for Walleyes’ migration from Lake Erie's western and central basins to the eastern basin or to Lake Huron has not been conclusively established. Wang et al. (2007) hypothesized that Walleye migration stems from a search for cooler temperatures or greater prey availability. Given reductions in prey availability in Lake Huron (Riley et al. 2008), including a major collapse in the Alewife population (Dunlop and Riley 2013), migration rates could be in a state of flux if they stem from prey availability. The greater contribution of age-5+ Walleyes relative to age-3 and age-4 fish during the summer months suggests that migration could be in response to temperature, with fish returning to Lake Erie in anticipation of the spawning season. One potential factor contributing to longer Saginaw Bay residence by younger Walleyes is that the sexually mature fraction is smaller for this group, and immature individuals are perhaps less compelled to return to Lake Erie as the spawning season approaches. Estimated age at 50% maturity for Walleyes in the western basin of Lake Erie is approximately 2.5 years for females and 1.0–2.5 years for males depending on the sampling method or agency (Wang et al. 2009). Therefore, differences in sexual maturity are unlikely to provide the full explanation, as our younger age-group consisted entirely of age-3 and age-4 Walleyes, most of which would be mature.
Fisheries management decisions for mobile species are complicated when fish traverse jurisdictional boundaries. In the Great Lakes region, management decisions are largely coordinated and cooperative among the U.S. states, Canadian provinces, and tribal/First Nation authorities bordering each of the lakes (Dochoda and Jones 2002; Gaden et al. 2013). Due to this history of cooperation and coordination, fishery managers in the Great Lakes region may be more adept or better prepared for dealing with management of the Walleye and other species that have a tendency to move large distances. Nevertheless, the exchange of individuals among Lakes Erie, St. Clair, and Huron complicates an already complex management system, and it may be necessary to hold discussions about how the interests of the respective parties will be represented in situations of fish migrating between lakes. We believe such discussions would be beneficial given that other studies have documented cases of fish migration between Great Lakes (Adlerstein et al. 2007; Ebener et al. 2010), and a lack of understanding as to the extent of interlake movement by fishes in the Great Lakes has been identified as a major information gap for managers (Landsman et al. 2011).
ACKNOWLEDGMENTS
This research was partially funded by the Great Lakes Fishery Trust (Project 2009.1080). Additional funding was provided by MDNR and by contributing partners of the Quantitative Fisheries Center at Michigan State University (MSU). We thank A. Cook (Ontario Ministry of Natural Resources), M. Thomas (MDNR), C. Schelb (MDNR), T. Kolb (MDNR), and C. Radek for their assistance in the laboratory or field. We additionally thank two anonymous reviewers for suggesting changes that improved this manuscript. This is publication 2015-09 of the MSU Quantitative Fisheries Center.




