A distinctive single-stranded DNA-binding protein from the Archaeon Sulfolobus solfataricus
Summary
Single-stranded DNA binding proteins (SSBs) have been identified in all three domains of life. Here, we report the identification of a novel crenarchaeal SSB protein that is distinctly different from its euryar-chaeal counterparts. Rather than comprising four DNA-binding domains and a zinc-finger motif within a single polypeptide of 645 amino acids, as for Methanococcus jannaschii, the Sulfolobus solfataricus SSB protein (SsoSSB) has a single DNA-binding domain in a polypeptide of just 148 amino acids with a eubacterial-like acidic C-terminus. SsoSSB protein was purified to homogeneity and found to form tetramers in solution, suggesting a quaternary structure analogous to that of E. coli SSB protein, despite possessing DNA-binding domains more similar to those of eukaryotic Replication Protein A (RPA). We demonstrate distributive binding of SsoSSB to ssDNA at high temperature with an apparent site size of approximately five nucleotides (nt) per monomer. Additionally, the protein is functional both in vitro and in vivo, stimulating RecA protein-mediated DNA strand-exchange and rescuing the ssb-1 lethal mutation of E. coli respectively. We dis-cuss possible evolutionary relationships amongst the various members of the SSB/RPA family.
Introduction
Archaea are a separate group of organisms distin-guished from the eubacteria through 16S rDNA sequence analysis. These prokaryotes are further subdivided into three diverse groups named the crenarchaeota, the euryarchaeota and the korarchaeota (Woese et al., 1990; Woese and Fox, 1977; Barns et al., 1996). Only members of the crenarchaeal and euryarchaeal groups, however, have been cultivated. Genomic studies suggest a significant evolutionary division between metabolic and informational processes in archaea. Whereas most intermediary metabolic processes strongly resemble those observed in eubacteria, genomic informational processes are generally thought to be more closely related to those found in eukaryotes (reviewed in Doolittle and Logsdon, 1998). Archaea utilize eukaryotic B-type DNA polymerases for replication, and their ribo-somal proteins, as well as translation initiation factors, are remarkably eukaryotic. The recent identification of archaeal snoRNA genes reveals an unexpected eukaryotic connection (Omer et al., 2000). Transcription also involves eukaryotic protein homologues, but the dis-covery of multiple TBP and TFB proteins in halophiles hints at a unique archaeal transcription mechanism (Baliga et al., 2000). Examination of archaeal recombination proteins suggests a definite similarity with eukaryotes. The archaeal DNA strand exchange protein, RadA, is more similar to its eukaryotic counterpart, Rad51 protein, than to the eubacterial RecA protein, both at the amino acid level (Sandler et al., 1999) and at the biochemical level (Seitz et al., 1998). The associated RadB protein is proposed to serve as a simpler archaeal version of eukaryotic Rad55/57 protein (Rashid et al., 1996; DiRuggiero et al., 1999; Komori et al., 2000), and archaeal Holliday junction resolvase protein char-acterization suggests they may also be eukaryotic in nature (Komori et al., 1999; Kvaratskhelia and White, 2000).
Single-stranded DNA (ssDNA) binding proteins (SSBs) are essential in most intracellular interactions that involve DNA, including replication, repair and recombination (Kowalczykowski et al., 1994; Lohman and Ferrari, 1994). Homologues of this class of proteins were identified in all three domains of life, as well as in viral genomes (Kowalczykowski et al., 1981; Lohman and Ferrari, 1994; Wold, 1997; Chedin et al., 1998; Kelly et al., 1998; Iftode et al., 1999). Despite the lack of strong homology at the amino acid level, preservation of both structural and domain organization suggests that SSBs are derived from a common evolutionary ancestor (Pfuetzner et al., 1997; Chedin et al., 1998; Raghunathan et al., 2000). While functionally equivalent, eubacterial SSB and the eukaryotic version, RPA, have distinctly different quaternary structures. In eubacteria, SSB is encoded by a single gene and the active form of the protein is a homotetramer in which each monomer provides one ssDNA-binding domain (Sancar et al., 1981; Lohman and Ferrari, 1994). In eukaryotes, the RPA complex from both humans and yeast is composed of three distinct subunits that together provide a total of four ssDNA-binding domains (Brill and Bastin-Shanower, 1998). Recent descriptions of archaeal SSB homologues from the euryarchaeal branch of the archaeal domain demonstrate their amino acid sequences are more similar to eukaryotic RPA than to eubacterial SSB (Chedin et al., 1998; Kelly et al., 1998). However, these archaeal proteins maintain multiple ssDNA-binding domains within one or just a pair of polypeptides and, therefore, are expected to function as monomers or heterodimers rather than as a homotetramer (as does E. coli SSB). It was proposed that these archaeal RPA homologues are evolutionarily related to eukaryotic RPA through gene duplication and recombination events (Chedin et al., 1998).
Genome sequencing of several archaeons simplified molecular analysis in these organisms. Whereas a number of euryarchaeal genome sequences have been determined, to date, the only publicly available crenarchaeal genome sequences are that of Aeropyrum pernix (Kawarabayasi et al., 1999) and Sulfolobus solfataricus (She et al., 2001). Here we describe the identification of a novel SSB with distinct genetic organization and sequence from S. solfataricus as well as a closely related open reading frame from A. pernix. To demonstrate that the proteins encoded by the identified open reading frames represent true SSB homologues, we heterologously expressed and purified S. solfataricus SSB protein, and we investigated its structure and ssDNA-binding activity, its ability to complement a eubacterial SSB mutation in vivo, and its effect on DNA strand exchange.
Results
Crenarchaeal homologues of ssDNA binding proteins
Previous searches of genome sequences identified open reading frames (ORFs) with homology to human RPA70 from the euryarchaeons Methanococcus jannachii, Meth-anobacterium thermoautrophicum and Archaeoglobus fulgidus (Chedin et al., 1998; Kelly et al., 1998). Each of these sequences consists of multiple ssDNA-binding domains contained in one or a pair of ORFs, which share a significant degree of sequence homology among themselves. In one case, the protein encoded by this homologous sequence was shown to be an RPA homologue, both structurally and functionally (Kelly et al., 1998). However, there has been no corresponding homologue of RPA identified in members of the other branch of the archaeal domain, the crenarchaea. The availability of sequence information from both Aeropyrum pernix (Kawarabayasi et al. (1999) and Sulfolobus solfataricus (She et al., 2001) permitted a search for ssDNA-binding protein sequences in the crenarchaea.
A survey of genome sequences from A. pernix and S. solfataricus with the program BLAST (Altschul et al., 1990) revealed a single small ORF in each genome with sequence similarity to MJ1159, the RPA homologue from M. jannaschii. Both ORFs code for proteins that are strikingly similar to the first ssDNA-binding domain of the M. jannaschii RPA (Fig. 1A), but are much shorter than the M. jannaschii protein. The S. solfataricus sequence (SSO2364) is 477 basepairs in length and encodes a protein of 148 amino acids whereas the A. pernix sequence (gi5105001) is 429 basepairs in length and encodes a protein of 143 amino acids; in contrast, the M. jannaschii protein is 645 amino acids in length. The S. solfataricus protein has a predicted pI of 9.0 and the A. pernix protein also has a predicted pI of 9.0. For comparison, the pIs of the T4 phage SSB homologue gp32, E. coli SSB and M. jannaschii RPA are 4.8, 5.4 and 4.7 respectively. The S. solfataricus protein has 52% similarity and 26% identity with MJ1159 from amino acids 68–170 and the A. pernix protein has 52% similarity and 25% identity with MJ1159 from amino acids 70–173. The two residues conserved in all archaeal sequences correspond to amino acids involved in DNA binding in human RPA70 protein domain B (Thr 359 and Trp 361 in the hsRPA70 sequence) (Bochkarev et al., 1997) (Fig. 1A). Additionally, the S. solfataricus sequence shares two other amino acids with human RPA70 that are implicated in DNA binding, Phe 386 and Ser 396 (Bochkarev et al., 1997).

Alignment of ssDNA-binding domain protein sequences. Crenarchaeal sequences are aligned with the first ssDNA-binding domains of M. jannachii RPA, S. cerevisiae RPA70 and H. sapiens RPA70. Identical residues are shaded black and conserved residues are shaded grey. The asterisk symbol (*) indicates residues identified in the H. sapiens RPA70 that interact with DNA (Bochkarev et al., 1997), whereas an ‘x’ indicates residues that are identical between the two crenarchaeal sequences. The consensus is shown beneath the alignment. Sequence accession nos. are as follows: S. solfataricus SSB, SSO2364; A. pernix SSB, gi5105001; M. jannachii RPA, MJ1159; S. cerevisiae RPA70, gi6319321; H. sapiens RPA70, gi1350579.
The two crenarchaeal protein sequences show a remarkable level of homology between each other, exhibiting 69% similarity and 47% identity (Fig. 1). Both of the newly identified ORFs are significantly shorter than their euryarchaeal and eukaryotic counterparts, each comprising only a single ssDNA-binding region as opposed to closely related tandem repeats (four, in the case of the M. jannaschii protein) of multiple binding regions in a larger polypeptide. Examination of nearby regions in each genome revealed no further sequences that might encode for portions of a ssDNA-binding protein, suggesting either that S. solfataricus SSO2364 and A. pernix gi5105001 are the only genes responsible for producing SSB for these crenarchaeons or that there were other genes elsewhere in the genome that encoded the remainder of the subunits for an RPA-like protein. No other sequence with significant similarity to MJ1159 is apparent elsewhere in the complete genomes of either A. pernix or S. solfataricus, suggesting that these proteins may serve as the sole SSBs in these organisms.
The crenarchaeal proteins structurally resemble E. coli SSB protein
In contrast to other archaeal SSB homologues that resemble eukaryotic RPAs, it appears that the overall architecture of the crenarchaeal versions is distinctly different. Euryarchaeal RPA homologues are comprised of multiple DNA-binding domains, within one or a pair of proteins, and presumably function in monomeric or heterodimeric forms respectively (Fig. 2). The ORFs from both A. pernix and S. solfataricus, however, are distinguished by a single core ssDNA-binding domain of about 100 amino acids in length, identical to that observed in eubacterial ssDNA-binding proteins. This suggests that the quaternary structure of the crenarchaeal SSB protein might be analogous to the eubacterial version, and could be tetrameric. Additionally, the crenarchaeal proteins lack the C-terminal zinc finger of the euryarchaeal RPAs and, instead, contain a number of acidic residues in this region. This is analogous to the C-terminal structure of E. coli SSB, which is required for protein–protein interactions (Williams et al., 1983; Chase et al., 1985; Kelman et al., 1998). Also absent in the crenarchaeal sequences is an approximately 100-residue N-terminal region found in euryarchaeal RPAs that is believed to be involved in protein binding (Wold, 1997).
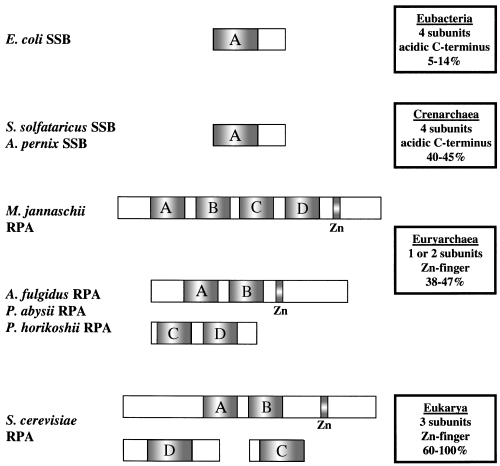
Schematic representation of the domainal architecture of ssDNA-binding proteins in all three domains of life. Homologous DNA-binding domains are represented by shaded boxes and the location of the zinc-finger motif is indicated. A summary of the attributes of each type of ssDNA-binding protein is represented in the boxes. A range of percentage similarities for proteins from each domain to the first single-stranded DNA-binding domain of S. cerevisiae RPA70 was determined using the BestFit program and is indicated.
The SSB protein homologue of phage T4, gp32, has little sequence homology to bacterial SSBs but displays both structural similarity with these proteins and an acidic C-terminus (Shamoo et al., 1995). Much like gp32, how-ever, the crenarchaeal SSBs share minimal sequence homology with E. coli SSB. BestFit alignment of the crenarchaeal sequences with E. coli SSB shows 35% similarity and 31% identity over only 48 amino acids for SsoSSB, and 43% similarity and 38% identity over only 60 amino acids for ApeSSB.
Expression and purification of the SSB homologue from S. solfataricus
To determine if SSO2364 from S. solfataricus did indeed encode a functional SSB protein homologue, we purified and characterized the candidate protein. The SSO2364 ORF was cloned into the pET21a bacterial expression vector and subsequently heterologously expressed in E. coli. The bacterial strain carried a plasmid encoding three rare tRNA genes in an effort to reduce translational pausing, as the predicted protein sequence contains 11 codons that are rare in E. coli, four of which are present in the final 50 bases of the ORF. The expressed protein is soluble and was purified to near homogeneity by first heat-treating the cell sonicate at 80°C to denature all mesophilic proteins. This step was followed by affinity chromatography on ssDNA-cellulose, and by anion ex-change chromatography on Resource Q. The recombinant protein eluted from ssDNA-cellulose at 0.75 M NaCl, which is identical to the salt concentration necessary for elution of E. coli SSB (Lohman et al., 1986). In contrast, elution of either yeast or M. jannachii RPA from the same matrix requires 1.5 M NaCl with the addition of 40% ethylene glycol, 1.3 M potassium thiocyanate or 1.5 M sodium thiocyanate (Heyer et al., 1990;Henricksen and Wold, 1994; Kelly et al., 1998), suggesting that the S. solfataricus SSB (SsoSSB) protein is chemically more similar to the bacterial protein than it is to RPA. The SsoSSB protein eluted at approximately 60 mM NaCl from Resource Q and pooled fractions yielded essentially pure protein (greater than 98% purity based on Coomassie brilliant blue-stained gels). Examination of the purified protein by SDS/PAGE revealed a single band with a molecular weight of approximately 16 kDa, in close agreement with the predicted molecular weight of 16 138 Da as determined from the amino acid sequence (Fig. 3).

SDS–PAGE of the purified SsoSSB protein. Samples were subjected to SDS–PAGE and gels were stained with Coomassie brilliant blue R250. The samples loaded were: uninduced crude cell sonicate (lane 1, 100 μg protein); induced crude cell sonicate (lane 2, 100 μg protein); heat-treated clarified sonicate (lane 3, 80 μg protein); pooled ssDNA-cellulose fractions (lane 4, 15 μg protein); and concentrated Resource Q fractions (lane 5, 15 μg protein). The arrow indicates the position of the SsoSSB protein.
SsoSSB protein forms tetramers in solution
As E. coli SSB protein assembles into a tetramer, it was consequently of interest to determine if the purified S. solfataricus protein would multimerize in solution. Analytical gel filtration was used to determine the native form of the recombinant protein. Examination of the SsoSSB protein in solution revealed that it was present in three distinct species: monomeric, dimeric and tetrameric forms, at 18 kDa, 36 kDa and 62 kDa respectively (Fig. 4A). As demonstrated by a representative elution profile (Fig. 4B), the composition of the purified material was primarily tetrameric, whereas somewhat less dimeric protein was present. A significantly smaller quantity of the monomeric form of SsoSSB protein was observed. This result indicates that the SsoSSB protein can indeed form tetramers in solution. As might have been expected, the M. jannachii RPA protein does not multimerize in solution (Kelly et al., 1998; unpublished observation).
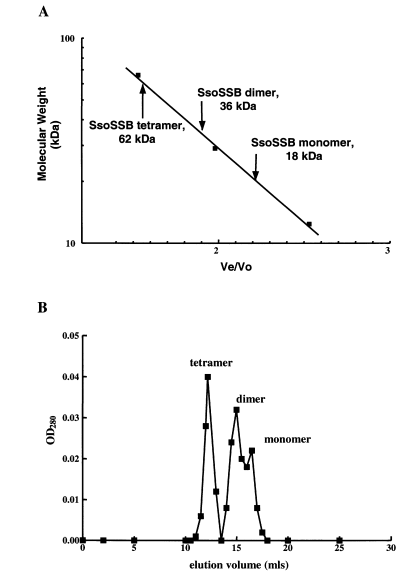
Gel filtration chromatography of SsoSSB protein. A. Elution of purified SsoSSB protein relative to molecular weight standards: BSA (66 kDa), carbonic anhydrase (29 kDa) and cytochrome c (12.4 kDa), which are represented by closed squares (▪). Ve/Vo, elution volume divided by void volume. B. A representative elution profile for purified SsoSSB protein. Closed squares (▪) represent optical density at 280 nm (OD280) for the elution volume indicated.
SsoSSB protein binds ssDNA
To determine the occluded binding site size of the protein and to evaluate the activity of the SsoSSB protein, we performed acrylamide gel mobility-shift assays with a radioactively labelled single-stranded oligonucleotide that was 63 nucleotides in length (Fig. 5). When a fixed quantity of radiolabelled oligonucleotide was incubated with increasing quantities of protein, a band of reduced mobility was observed. The apparent mobility further decreased upon addition of more protein, finally achieving a constant mobility after the addition of 2 μM protein. The absence of discrete species implied rapid equilibration between bound and free forms of the complexes and, consistent with this possibility, more discrete species were visualized when electrophoresis was performed more rapidly (at high voltage; data not shown). The addition of linear pUC19 DNA to a 100-fold molar (nucleotide) did not alter the mobility-shift pattern, demonstrating that the SsoSSB has an, at least, 100-fold greater affinity for ssDNA than for dsDNA (data not shown). Saturation was achieved at a ratio of approximately one SSB protein molecule to five nucleotides. The M. jannaschii RPA protein has a site size of 15–20 nucleotides (Kelly et al., 1998) whereas the site size of E. coli SSB protein, depending on solution conditions, varies from 8–16 nucleotides per monomer. The apparent site size of SsoSSB protein is similar to the seven-nucleotide site size observed for phage T4 SSB protein, gp32. Binding of SsoSSB protein to ssDNA does not appear to be a co-operative process under these conditions, as intermediately shifted species are evident instead of the approximately two-state transitions that typify co-operative binding. Rather, a steady reduction in the mobility of protein-DNA complexes occurs as protein concentration is increased, suggesting that binding of SsoSSB protein to ssDNA is distributive in nature. The observed band retardation pattern is probably the result of a combination of this non-co-operative binding and rapid equilibration of protein–DNA complexes during electrophoresis.
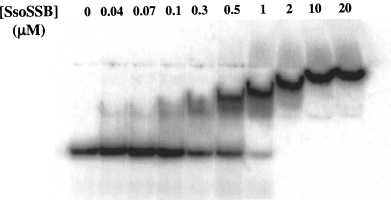
Gel mobility-shift analysis of SsoSSB protein binding to ssDNA. Increasing concentrations of protein (0.04 to 20 μM) were added to a constant concentration of 63-mer oligonucleotide (10 μM nucleotides).
SsoSSB protein is a functional SSB homologue
In E. coli, SSB is an essential protein. A number of mutations were used to elucidate the function of E. coli SSB protein; one mutation is the temperature-sensitive mutation called ssb-1. The ssb-1 mutation is an alteration of amino acid 55 from a histidine to a tyrosine, which is believed to destabilize the tetrameric SSB protein complex upon shifting temperature from 30°C to the non-permissive temperature of 43°C, resulting in a lethal phenotype (Chase et al., 1983). Overexpression of wild-type E. coli SSB protein encoded on a plasmid can overcome the lethality of the mutation (Chase et al., 1983) as can the SSB protein of bacteriophage P1 (Lehnherr et al., 1999). To test the in vivo functionality of SsoSSB, the protein was overexpressed in the E. coli strain KLC789 that carries the ssb-1 allele. The SsoSSB plasmid was transformed into KLC789 along with a plasmid carrying an arabinose-inducible T7 polymerase gene. Cells were grown in the presence of arabinose for 16 h at the permissive temperature to allow overexpression of the SsoSSB protein, whereas control cultures were cultivated in the presence of glucose. Cells were then shifted to the non-permissive temperature and monitored for further growth (Fig. 6A). The presence of the ORF encoding SsoSSB protein in pET21a permitted continued growth of the ssb-1 mutant strain whereas the pET21a vector alone was unable to rescue the lethal phenotype. Rescued growth required the presence of arabinose, as control experiments with glucose showed no increase in optical density that would be consistent with continued growth. To verify that the cells containing the SsoSSB plasmid were indeed still growing, viable counts for each time-point were determined by plating (Fig. 6B). Following the temperature shift, the control cultures continued to grow for a brief period and then dramatically lost viability. In contrast, the cells containing the SsoSSB plasmid displayed a continuous increase in viable counts. These results indicate that SsoSSB protein is capable of replacing E. coli SSB protein in vivo and that it functions at 43°C.

Overexpression of SsoSSB protein rescues the lethal phenotype of an E. coli ssb-1 mutation. KLC789 cells containing the pTara arabinose-inducible T7 expression plasmid, and either pET21a (circles) or the SsoSSB expression vector (squares) were grown at 30°C in either arabinose (closed symbols) or glucose (open symbols). Cultures were shifted to 43°C at the point indicated by the arrow. A. Optical densities were monitored spectrophotometrically at 600 nm. B. Colony forming units (cfu) were determined by plating in triplicate and the points shown are averages of the replicates.
SsoSSB protein stimulated DNA strand exchange by E. coli RecA protein
It was shown previously that heterologously expressed SSB proteins can stimulate DNA strand exchange mediated by E. coli RecA protein (Egner et al., 1987). To demonstrate the functionality of the SsoSSB protein in at least one nucleic acid metabolic function in vitro, DNA strand exchange reactions were performed (Fig. 7). Purified SsoSSB protein was capable of substituting for E. coli SSB protein in DNA strand exchange reactions mediated by E. coli RecA protein at 37°C using homologous φX174 ss- and dsDNA. Clear enhancement of DNA strand ex-change, as determined by production of nicked circular product, was observed with SsoSSB protein (Fig. 7, lane 3). Interestingly, the M. jannaschii RPA protein can also promote RecA-mediated DNA-strand exchange (E.M. Seitz, C.A. Haseltine and S.C. Kowalczykowski, unpublished observations). The reduced amount of nicked circular product in reactions containing SsoSSB protein relative to levels with E. coli SSB protein may be the result of performing the experiment at temperatures at which SsoSSB protein is not as effective. The optimal temperature for the SsoSSB protein is expected to be near the growth temperature of S. solfataricus, which is 80°C. As yet, no enhancement of strand exchange activity through addition of SsoSSB protein to S. solfataricus RadA protein-containing reactions at high temperature has been observed (unpublished observations), though this may be a consequence of inappropriate assay conditions or may signify a postsynaptic role for SsoSSB protein at physiological temperatures.

SsoSSB protein stimulates DNA strand exchange by E. coli RecA protein. Lane 1, no protein; lane 2, RecA protein; lane 3, RecA protein and SsoSSB protein; lane 4, RecA protein and E. coli SSB protein. The abbreviations are: JM, joint molecules; NC, nicked circular dsDNA; DS, dsDNA; and SS, ssDNA. A schematic representation of the formation of nicked circular dsDNA and joint molecules from starting substrates is shown above the gel.
Discussion
Divergence between the archaeal and eukaryotic lineages is more recent than the divergence of the bacte-rial and eukaryotic/archaeal groups (Olsen and Woese, 1997). Accordingly, a number of features shared by eukaryotes and archaea, but not bacteria, including replication and transcription proteins were probably obtained after evolutionary divergence of these three groups. In contrast, features found in archaea and bacteria, but not in eukaryotes, including morphological attributes (a lack of organelles and nucleus), coupling of transcription and translation, conjugative mechanisms and a single circular genome may be reminiscent of a more ancient, shared ancestor.
However, despite the logic of this viewpoint, the evolutionary behaviour of the archaea is not so simplistic. Our results indicate that, in contrast to the canonical expectation that all archaeal ssDNA binding proteins would be RPA-like, the crenarchaeal SSBs share sequence homology with eukaryal RPAs but structural homology with bacterial SSBs. Both E. coli SSB and SsoSSB proteins elute from ssDNA-cellulose at the same salt concentration, and both form tetramers in solution. The apparent binding site size is approximately five nucleotides, which is consistent with the site size observed for T4 g32 protein and the low site size binding mode of E. coli SSB protein, but is one quarter that of M. jannachii RPA protein, which has four DNA-binding domains. Another feature that makes the crenarchaeal SSB protein more eubacterial is that SsoSSB protein and ApeSSB protein are missing the zinc finger motif near the C-terminus, which is present in both euryarchaeal and eukaryal RPAs, suggesting that this motif was acquired after the separation of the two archaeal branches. Furthermore, the acidic residues in the SsoSSB protein C-terminal region are similar to those found in bacterial proteins and may have been maintained from the last shared bacterial and archaeal ancestor. SsoSSB protein resembles E. coli SSB chemically in that they both elute from ssDNA-cellulose at 0.75 M NaCl whereas the euryarchaeal and eukaryal RPAs require higher salt concentrations and the addition of ethylene glycol, potassium thiocyanate or sodium thiocyanate for elution from this matrix. Finally, SsoSSB protein can functionally substitute for E. coli SSB protein both in vivo and in vitro. In vivo, overexpression of the SsoSSB protein can overcome the lethal ssb-1 temperature-sensitive mutation in E. coli; in vitro, the SsoSSB protein can replace the E. coli SSB protein in DNA strand exchange reactions mediated by RecA protein.
Concurrent with our efforts, the same crenarchaeal protein was identified by another laboratory using biochemical criteria (Wadsworth and White, 2001). The DNA-binding characteristics of SsoSSB protein were found to be similar to those we describe here. No apparent multimerization of the protein was demonstrated, in contrast to our observation. In our experience, however, heating of the SsoSSB protein was necessary for formation of the tetrameric structure as observed by gel filtration and for activity in DNA strand-exchange. The absence of this preheating step by Wadsworth and White could account for this seemingly minor discrepancy.
Overall, the archaeal ssDNA binding proteins show more sequence similarity to eukaryotic RPA proteins than to E. coli SSB protein. However, the common sequences and DNA-binding domain utilization hint at a conserved evolutionary relationship between RPA and SSB (Fig. 2). Strong homology among the archaeal proteins suggests a common evolutionary origin, and the crenarchaeal SSB proteins may represent a link to ancient ssDNA-binding proteins. The crenarchaeal SSB protein, being one of the simplest in structure, may represent the earliest form of single-stranded DNA-binding proteins involved in recombination, and it may serve as a model for understanding structure, function and evolution of the conserved core ssDNA-binding domain.
Crenarchaea and euryarchaea are distinguished from each other not only by single-stranded DNA-binding proteins as described here, but also by double-stranded DNA-binding proteins. Whereas eukaryotic-like histone proteins have been identified and extensively studied in members of the euryarchaea (Sandman and Reeve, 2000), no such homologues are apparent in members of the crenarchaea. Instead, crenarchaea maintain sequences that code for small, basic dsDNA-binding proteins (Agback et al., 1998; Faguy and Doolittle, 1999). These proteins are proposed to be similar to histone-like proteins in eubacteria, especially Sso7d, which was shown to be analogous to the dsDNA-binding protein HU (Lopez-Garcia et al., 1998). It appears that the crenarchaea have maintained both single-stranded and double-stranded DNA-binding proteins that are more similar to those found in bacteria than they are to eukaryotic homologues. Crenarchaeal dsDNA-binding proteins and SSB may have evolved separately from mechanisms employed by eukaryotes or, alternatively, crenarchaea diverged earlier, before the co-evolution of eukaryotic-like histones and RPA. The use of a eubacterial-like SSB may necessitate a eubacterial DNA-binding protein for the compaction of DNA. The discovery of an SSB homologue in the crenarchaea that is significantly more similar to eubacterial SSB, both structurally and biochemically, than it is to either euryarchaeal or eukaryotic RPA further establishes the evolutionary distance between the two archaeal phyla.
Experimental procedures
Alignment of protein sequences
The Sulfolobus solfataricus ssb sequence (SSO2364) encoding the SSB protein (SsoSSB) was identified using BLASTP at the S. solfataricus genome website: http://www-archbac. upsud.fr/projects/sulfolobus. The Aeropyrum pernix ssb sequence (gi5105001) encoding the SSB protein (ApeSSB) was identified using BLAST at the PEDANT website: http://pedant.gsf.de/. Both ORFs were recognized through their homology to MJ1159, encoding the Methanococcus jannaschii RPA protein (Chedin et al., 1998). Subsequent alignments were performed using the ALIGN program at http://www.toulouse.inra.fr/multalin.html and additional features were highlighted by manual adjustment. BestFit comparisons were performed using the Wisconsin Package Version 10.1, Genetics Computer Group (GCG), Madison, Wisconsin and were between S. cerevisiae RPA70 single-stranded DNA-binding region 1 (gi6319321, amino acids 301–399) and the following single-stranded DNA-binding protein sequences; Archaeoglobus fulgidus (gi11497994), A. pernix (gi5105001), Bacillus subtilis (gi2127217), Escheri-chia coli (gi134913), Homo sapiens (gi1350579), M. jannaschii (MJ1159), Methanobacterium thermoautotrophicum (gi2622495), Pyrococcus abyssii (gi5457718), Pyrococcus horikoshii (gi3258332), S. solfataricus (SSO2364), as well as between E. coli SSB protein (gi134913) and S. solfataricus (SSO2364). The A. fulgidus sequence used was one of two identified as homologous to MJ1159 and is the most homologous to the N-terminus of the M. jannaschii protein (Chedin et al., 1998). The M. thermoautotrophicum sequence was adjusted to account for the frameshift identified by Chedin and colleagues (Chedin et al., 1998).
Strains and cultivation
Sulfolobus solfataricus strain P2 (DSM 1616 (Zillig et al., 1980)) was the generous gift of Dennis Grogan (University of Cincinnati) and was grown at 80°C as described (Rolfsmeier et al., 1998) at a pH of 3.0 in screw-cap flasks as described (Rolfsmeier and Blum, 1995). Basal salts medium was Allen’s medium (Allen, 1959), as modified by Brock (Brock et al., 1972), and was supplemented with tryptone to a final concentration of 0.2% (w/v). Growth was monitored spectrophotometrically at a wavelength of 540 nm. Escherichia coli strains were DH5α (φ80 dlacZΔM15, endA1, recA1, hsdR17 (rk−,mk+), supE44, thi-1, gyrA96, relA1, Δ(lacZYA-argF)U169); BL21(DE3) (ompT [lon] hsdSB (rB−mB−; an E. coli B strain) with DE3, a λ prophage carrying the T7 RNA polymerase gene); and KLC789 (F−, metA7, rha8, thyA36, amp50, deoC2, ssb-1) (Chase et al., 1983) from laboratory collections or BL21(DE3) CodonPlus ultracompetent cells from Stratagene. Escherichia coli was propagated in Luria–Bertani (LB) medium (Sambrook et al., 1989) at 30°C in Ehrlenmeyer flasks shaken at 250 r.p.m.
PCR amplification and cloning of the SsoSSB gene
Genomic DNA was prepared from S. solfataricus cells as described previously (Rolfsmeier et al., 1998). Polymerase chain reaction (PCR) was performed using 10 mM potassium chloride, 10 mM ammonium sulphate, 2 mM magnesium chloride, 20 mM Tris-Cl (pH 8.75), 0.1% Triton X-100, 100 μM dNTPs, 100 pmol primers, 2 ng template DNA and 2.5 U of recombinant Pfu DNA polymerase (Stratagene). The primers for amplification of SsoSSB were: 5′-CGGGATCCCCTTTCA TTAACACATAGATTTATAAATGG-3′ (SSB-F) and 5′-CGGGA TCCGGAGCAAGCTCGTATACTTTGTCTCTAGCC-3′ (SSB-R). All primer sequences were chosen based on sequence information presented at the S. solfataricus genome website: http://www-archbac.u-psud.fr/projects/sulfolobus. PCR was performed using a 55°C annealing temperature and the resulting PCR products were digested with BamHI and ligated into the BamHI site of pUC19. Ligated molecules were transformed into DH5α as described previously (Rolfsmeier et al., 1998). Plasmids from transformants were isolated using the Qiagen midiprep system and DNA sequences were determined using BigDye dRhodamine Terminator chemistry (Perkin-Elmer) at the Division of Biological Sciences Automated DNA sequencing facility at UC Davis.
Overexpression of SsoSSB protein
Sequence information obtained from the pUC19 clones was used to design a forward PCR primer with an NdeI site at the starting ATG codon for SsoSSB. The gene sequence was re-amplified from the cloned template using the new forward primer (5′-GTGAGTCGAGTCATATGGAAG-3′) and the original reverse primer. The resulting product was digested using NdeI and BamHI before ligation into pET21a (Novagen), which had been digested with the same enzymes to place the gene under the control of the T7 promoter. Ligation products were transformed into the CodonPlus strain (Stratagene) and transformants were cultivated at 30°C in LB containing 100 μg ml−1 of ampicillin until mid-log phase.
Purification of SsoSSB protein
BL21(DE3) CodonPlus cells (Stratagene) harbouring the pET21a SsoSSB expression construct were grown at 30°C in a 500 ml volume to an OD600 of 1.0. IPTG was added to a final concentration of 1 mM and expression was allowed to continue for 2 h. Cells were harvested by centrifugation and stored at –20°C until processing. The frozen cell pellet was resuspended in 4 ml of 10 mM Tris-Cl (pH 7.5), 1 mM EDTA (TE) with 50 mM NaCl and sonicated to disrupt the cells. The sonicate was heat treated at 80°C for 1 h, and insoluble material was removed by centrifugation. Clarified sonicate was applied to a ssDNA cellulose column equilibrated in 30 mM Tris-Cl (pH 7.5), 1 mM EDTA, 1 mM DTT and 10% glycerol; the column was washed with the same buffer containing 0.5 M NaCl and 0.75 M NaCl at room temperature. Fractions eluting at 0.75 M NaCl were pooled and dialysed into buffer containing 20 mM Tris-Cl (pH 7.5), 1 mM DTT, 1 mM EDTA and 10% glycerol. This material was then applied to a Resource Q column (Pharmacia) equilibrated with the same buffer at room temperature. Protein was eluted using a gradient of 50 mM NaCl to 1 M NaCl in the same buffer; the SsoSSB protein eluted from the column at approximately 60 mM NaCl. The protein was pooled and concentrated using dry polyethylene glycol and then dialysed against 25 mM Tris-HCl (pH 7.5), 20 mM NaCl, 1 mM EDTA, 1 mM DTT and 10% spectral grade glycerol, and stored at 4°C. Protein concentrations were obtained by spectrophotometric absorbance at a wavelength of 280 nm, using an extinction coefficient of 12 660 M−1 cm−1 as determined with the ProtParam tool at the ExPASy website (http://expasy.cbr.nrc.ca/ tools/protparam.html).
Gel filtration of SsoSSB protein
Fast protein liquid chromatography (FPLC) was performed at 4°C using a Superose 12 column (Pharmacia) and 25 mM Tris-HCl pH 7.5, 1 mM DTT, 100 mM NaCl and 1 mM EDTA as the running buffer. Molecular size standards were BSA (66 kDa), carbonic anhydrase (29 kDa) and cytochrome c (12.4 kDa) and were prepared in running buffer. A total of 10 μg of SsoSSB protein was loaded on the column in a volume of 100 μl. Elution profiles were determined by monitoring OD280 readings and a standard curve was prepared by plotting elution volume (Ve)/void volume(Vo) against the molecular mass of the size standards. The value of Vo was determined by elution of Dextran blue from the Superose 12 column.
Gel mobility-shift analysis
The 63-mer oligonucleotide 5′-ACAGCACCAATG AAATCTA TTAAGCTCCTCATCGTCCGCAAAAATATCGTCACCTCAA AAGGA-3′ was end-labelled with 32P using T4 polynucleotide kinase (NEB). SsoSSB protein was incubated at the indicated concentrations with 10 μM (nucleotides) of the 32P-labelled oligonucleotide for 30 min at 75°C in buffer containing 30 mM Tris-OAc (pH 7.5), 10 mM MgOAc2, 5 mM NaCl, 0.1 mM DTT and 50 μg ml−1 of BSA. Increasing concentrations of linearized pUC19 were used as the dsDNA competitor as indicated in the text. Loading dye was then added, and the samples were applied to a vertical 10% acrylamide gel prepared with 1× TBE buffer (0.089 M Tris-borate, 0.089 M boric acid, 0.002 M EDTA).
In vivo complementation
The SsoSSB expression vector or pET21a (empty vector) was transformed into E. coli strain KLC789 (Chase et al., 1983) containing pTara, a T7 polymerase expression vector that is inducible by arabinose addition. The pTara plasmid was the generous gift of Kathleen Mathews (Rice University) (Wycuff and Matthews, 2000). Transformants were propagated in LB medium lacking yeast extract with 0.2% (w/v) arabinose, 100 μg ml−1 of ampicillin and 30 μg ml−1 of chloramphenicol for 16 h at 30°C to allow phenotypic overexpression of SsoSSB protein. Control cultures were propagated identically, except 0.2% (w/v) glucose was substituted for arabinose. Cells were subcultured into fresh medium without chloramphenicol and grown at 30°C until they were shifted to the non-permissive temperature of 43°C. Optical densities were monitored spectrophotometrically at a wavelength of 600 nm. Colony forming units (cfu) per millilitre were determined by plating serial dilutions of each time-point in triplicate on LB medium. Plates were incubated overnight at 30°C before scoring for viable counts.
DNA strand exchange reactions
Escherichia coli RecA protein (11 μM) was incubated with φX174 ssDNA (New England Biolabs) at a concentration of 33 μM (nucleotides) in 30 mM Tris-OAc (pH 7.5), 10 mM DTT, 20 mM MgOAc, 2.5 mM ATP, and 5 μg ml−1 of BSA at 37°C for 10 min. After the addition of either 2.2 μM SsoSSB protein or E. coli SSB protein, reaction mixtures were incubated at 37°C for another 5 min before the introduction of PstI-linearized φX174 dsDNA (New England Biolabs) at a concentration of 33 μM (nucleotides). The reaction mixtures were then incubated at 37°C for 90 min and were stopped by the addition of SDS to a final concentration of 0.6% and proteinase K to a final concentration of 1 μg ml−1. Deproteinization of the reaction mixtures was carried out at 37°C for 10 min.
Gel electrophoresis
Agarose gels for evaluation of cloning procedures were prepared at 0.8% and run at approximately 150 V in TBE buffer before staining with ethidium bromide. Tricine SDS/PAGE, used to monitor protein purification, was prepared with a 4% stacking gel and a 20% separating gel as described (Price and Shand, 2000), and run at 100 V. Acrylamide gels for gel mobility-shift analysis were prepared at 10% in TBE buffer and run at 100 V for 3 h at 65°C before exposure to phosphorimaging screens and analysis with a Storm 840 PhosphorImager (Molecular Dynamics). Agarose gels for evaluation of DNA strand exchange products were prepared at 1% and run at approximately 30 V in TAE buffer (0.04 M Tris-OAc, 0.002 M EDTA) for 15 h before staining with ethidium bromide.
Acknowledgements
We thank the following members of our laboratory for their helpful discussions and critical comments on the manuscript: Andrei Alexeev, Piero Bianco, Carole Bornarth, Mark Dillingham, Noriko Kantake, Alex Mazin, Olga Mazina, Katsumi Morimatsu, Jim New, Erica Seitz, Maria Spies and Tomohiko Sugiyama. This work was supported by NSF Postdoctoral Fellowship in Microbial Biology #0074380 to C.A.H and NIH grant GM62653 to S.C.K.