Lack of genetic and plumage differentiation in the red-billed quelea Quelea quelea across a migratory divide in southern Africa
Abstract
A migratory divide usually signals the presence of a geographical region over which other traits, such as morphology and genotypes, also undergo rapid change. A migratory divide has been hypothesized in central southern Africa for the abundant migratory weaver, the red-billed quelea Quelea quelea. The positioning of the divide in the region is based on the patterns of rainfall in the region that stimulate the annual migrations of queleas. Evidence indicates that premigratory queleas near the divide show two distinct preferred directions for migration. We used eight polymorphic microsatellite loci and a range of plumage characters to determine whether there was population structure among red-billed queleas in southern Africa, and specifically whether this structure coincided with the location of the migratory divide. There was no evidence of population genetic structure. An amova revealed no significant differences between samples taken either side of the migratory divide. Similarly, there was no geographical variation in plumage patterns across southern Africa. For both microsatellites and plumage characteristics, the variation that does exist occurs within each sampled site, with little differentiation between sites. We were therefore unable to find any evidence that either plumage or microsatellite genotypes varied in a similar way to migratory direction preference in red-billed queleas in southern Africa. This is perhaps because the migratory divide does not act to separate individuals into populations within which genetic and plumage differentiation can be maintained.
Introduction
Birds are known to exhibit generally low levels of genetic differentiation because of their high dispersal ability (Crochet 1996; Webster et al. 2002). In general, species that disperse less readily are more likely to show increased differentiation at the subspecies level (Belliure et al. 2000). Despite this, long-distance migrants from across all taxa maintain population structure despite their dispersal abilities, because individuals exhibit different levels of site philopatry and differential mating on the breeding grounds [e.g. noctule bats Nyctalus noctula (Petit & Mayer 2000), various shorebirds (Haig et al. 1997) and beluga whales Delphinapterus leucas (O’Corry-Crowe et al. 1997)].
One way that population differentiation in migrants can be maintained even in a seemingly continuous population is through the presence of a migratory divide, where two populations showing different migratory directions meet (Chambelain et al. 2000). A migratory divide is essentially an abrupt change in a trait value (migration direction) along a geographical transect, and hence the same evolutionary processes responsible for the migratory divide could also give rise to clines in other trait values, such as morphological or genetic variation. For example, in central Europe blackcaps (Sylvia atricapilla) either side of a migratory divide migrate either southeast or southwest to reach Africa. The different migration directions are determined genetically (Helbig 1996). Similarly, the latitudinal migratory divide in central Sweden for the willow warbler (Phylloscopus trochilus) coincides with morphological and genetic differences (Bensch et al. 1999). Hence, even when there is apparent mixing of individuals, population structure can be maintained through the existence of a migratory divide, although it is perhaps inevitable that some hybridization does occur (Helbig et al. 2001).
In sub-Saharan Africa red-billed queleas Quelea quelea undertake seasonal long-distance migrations in response to the availability of their principal food, the seeds of annual grasses, which in turn is determined by seasonal patterns of rainfall (Ward 1971). Regional differences in rainfall patterns across Africa, and hence in migration pathways, have resulted in the segregation of quelea populations into five to six geographical zones, defining four currently recognized subspecies, based on variation in the breeding plumage of adult males (Ward 1971; Jones 1989a). Within these populations, however, there may exist further structure, based on the geographical position of breeding areas in the zone and the migration routes between them, which could keep groups of birds apart. In eastern Africa, for example, there is evidence of long-term cohesion within flocks and separation between them (Jaeger et al. 1986, 1989), suggesting a potential mechanism for population structuring. There have been indications of similar flock cohesion in southern Africa (Jones 1989b).
A more recent study of orientation among premigratory queleas in Zimbabwe pointed to the existence of a migratory divide in central southern Africa (Dallimer & Jones 2002). At the start of the rains, queleas have the option of migrating to early rains quarters to the northwest in Angola, or southeast in Mozambique and South Africa (Ward 1971; Jones 1989b). Such a migratory divide in the heart of the nonbreeding distribution has the potential to separate two populations of queleas as the breeding season begins and maintain population structure in an otherwise continuous migratory population (Fig. 1).

The timing and directions of red-billed quelea migrations in southern Africa. Contours mark the start of the wet season and the shaded area represents the range of quelea (Jones et al. 2000). In October (Oct) quelea move ahead of oncoming rain fronts, concentrating their numbers in the centre of the region. When these rainfronts meet, queleas migrate back over the rainfronts (Nov) either to the southeast or to the northwest creating a migratory divide. Quelea then begin breeding and moving back towards the centre of southern Africa (Feb).
Male queleas show conspicuous polymorphisms in breeding plumage (Dale 2000). Recording male plumage patterns has been advocated as a routine technique for monitoring quelea numbers and movements (Allan 1996) in order to infer relationships between breeding colonies (Jaeger et al. 1989; Manikowski et al. 1989) and to define subspecies (Ward 1966). Plumage patterns are inherited (Dale 2000) and appear to remain the same in a given area over a number of years (Manikowski et al. 1989). Although there are no data about the genetic control mechanisms that underlie these plumage polymorphisms, they could act as an indirect marker in the detection of a migratory divide and the assessment of migratory connectivity.
Molecular markers provide another indirect method of assessing the migratory connectivity of queleas in southern Africa. We used a series of polymorphic microsatellite markers to test the hypothesis that the migratory divide in central southern Africa coincides with a rapid change in genetic and plumage polymorphisms across a narrow geographical region. By combining both molecular and morphological data, we aim to investigate the role of a migratory divide in maintaining population structure and differentiation in an otherwise continuously distributed species.
Materials and methods
The migratory divide
Evidence for a migratory divide for red-billed quelea was found using orientation funnels on queleas of both sexes from a nonbreeding roost at Lake Manyame (17°49′ S 30°30′ E) on the Zimbabwean Highveld, a high plateau that runs down the spine of Zimbabwe (Dallimer & Jones 2002). Quelea aggregate in the nonbreeding season on the Zimbabwean Highveld (Jones 1989b) but are forced to leave the region at the onset of the rains as their grass seed food germinates. The patterns of rainfall in the region potentially offer two ‘escape routes’ for quelea spending the nonbreeding season on the Highveld, either northwest to Angola or southeast to South Africa and Mozambique (1, 2). The orientation of the divide, perpendicular to the two migratory directions taken by queleas in the orientation experiments, therefore appears to be c. 27°. We took samples of queleas from sites either to the northwest or southeast of this line. A few sample locations that were very close to the divide were not placed in either region and were excluded from analyses comparing queleas on opposite sides of the divide.
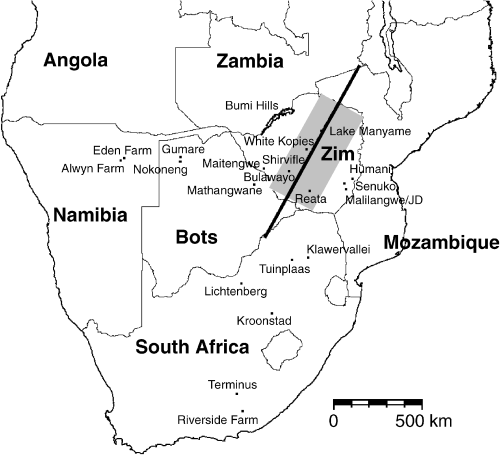
Sampling sites for red-billed queleas in southern Africa. The location of the hypothesized migratory divide is indicated with the thick black line. The shaded area represents the approximate extent of the Zimbabwean Highveld. Zim: Zimbabwe. Bots: Botswana.
Sampling
In total, 1397 male queleas in breeding plumage were sampled from 23 sites during 1998 and 1999 (Fig. 2, Table 1). All samples are from the range of the southern African subspecies Q. q. lathamii. Queleas are a serious agricultural pest and many governments actively control their impact on crops (e.g. Geertsema 1999). All sampling was carried out with the permission of government pest control agencies and most birds were collected dead following pest control operations. Each bird's plumage type was recorded, before a small (5 mm × 5 mm) piece of liver was removed. In some cases queleas were frozen until data gathering could be completed. Alternatively live queleas were caught in mist-nets, individuals were bled and their plumage patterns assessed. Blood samples were taken from the brachial vein on the underside of the wing using a sterile disposable hypodermic needle (12 mm × 0.4 mm) to puncture the vein. Blood was taken up using a 1-µL heparinized microhaematocrit capillary tube and immediately transferred to a 2-mL sample tube. The sample tubes contained a preservative consisting of 10 mm Tris HCl (pH 8), 100 mm EDTA (pH 8) and 2% w/v SDS. Once collected, samples were kept cool until being frozen for storage. Liver samples were stored in the same preservative.
| Site | Name | Region | Number of | |
|---|---|---|---|---|
| Genotypes | Plumage | |||
| AF | Alwyn Farm | North West | 45 | 53 |
| BU | Bulawayo | Central | 36 | 0 |
| ED | Eden Farm | North West | 44 | 43 |
| GU | Gumare | North West | 44 | 63 |
| HU | Humani | South East | 45 | 0 |
| JD | JDMalilangwe | South East | 35 | 0 |
| KR | Kroonstad | South East | 45 | 57 |
| KW | Klawervallei | South East | 41 | 62 |
| LM | Lake Manyame | Central | 45 | 0 |
| LT | Lichtenberg | South East | 44 | 70 |
| MA | Mathangwane | North West | 46 | 79 |
| NN | Nokoneng North | North West | 46 | 80 |
| NS | Nokoneng South | North West | 45 | 59 |
| RF | Riverside Farm | South East | 45 | 90 |
| RR | Reata | Central | 42 | 98 |
| SH | Shirville Farm | Central | 41 | 96 |
| TE | Terminus | South East | 45 | 92 |
| TU | Tuinplaas | South East | 44 | 55 |
| WK | White Kopjes | Central | 0 | 33 |
| XA | Senuko | South East | 48 | 88 |
| XB | Bumi Hills | North West | 46 | 97 |
| XC | Maitengwe | North West | 48 | 90 |
| XX | Malilangwe | South East | 48 | 92 |
| 968 | 1397 | |||
Population differentiation and subspeciation patterns in red-billed queleas have been defined previously on the basis of regional differences in male plumage colouration alone (Ward 1966). As a major aim of this study was to integrate information on plumage morphs with genetic variation to assess what degree of population structure might be associated with the migratory divide, we concentrated solely on males.
Molecular methods and data analysis
Eight polymorphic microsatellite loci were used in the population genetics analysis (Table 2). The eight loci were selected from a panel of 20 loci that amplified polymorphic products in the red-billed quelea (Dallimer 1999). The loci chosen from the initial panel were a selection of the least variable markers that amplified easily with no evidence of null alleles (Dallimer 2001). Details of the original species in which the loci were isolated, polymerase chain reaction (PCR) conditions and DNA extraction have been given elsewhere (Dallimer 1999). In all, 968 queleas from 22 sites were genotyped; up to 51 males were typed for each site (Table 1).
| Name | n | Allele size range (bp) | No. alleles | H O |
|---|---|---|---|---|
| Esc4 | 968 | 146–188 | 22 | 0.865 |
| Hru5 | 968 | 108–143 | 27 | 0.86 |
| Mcyu4 | 968 | 138–192 | 29 | 0.824 |
| Phtr2 | 968 | 107–143 | 16 | 0.754 |
| Phtr3 | 968 | 126–182 | 29 | 0.866 |
| WBSW1 | 968 | 141–213 | 29 | 0.787 |
| WBSW2 | 968 | 201–257 | 28 | 0.81 |
| WBSW4 | 968 | 131–193 | 31 | 0.878 |
Observed and expected heterozygosity estimates were calculated using genetix (Belkhir 1999). Deviations from Hardy–Weinberg expectations were tested using genepop 3.1d (Raymond & Rousset 1995). An exact Hardy–Weinberg test (Guo & Thompson 1992) based on a Markov chain algorithm was used to obtain an unbiased estimate of the exact probability of incorrectly rejecting the null hypothesis of Hardy–Weinberg equilibrium. The Markov chain was set to 10 000 steps with 1000 steps of dememorization. The algorithm was performed in 500 batches of 1000 iterations per batch. Tests for linkage disequilibrium between locus genotypes were performed in genepop under the null hypothesis that genotypes at one locus were independent from genotypes at the other locus. An exact test was performed together with a Markov chain process, as described above, to evaluate statistical significance (Guo & Thompson 1992). An exact test of population differentiation was carried out in genepop using the process outlined above to evaluate statistical significance. Sequential Bonferroni corrections (Rice 1989) were used in all cases with multiple tests.
θ was used as an estimator of pairwise FST (Weir & Cockerham 1984), as it is the least biased and most widely used statistic, and is believed to be more appropriate in situations with the number of loci available in this study (Gaggiotti et al. 1999). θ was estimated using the software package arlequin 2.0 (Schneider et al. 2000) between all site pairs. To test the significance of the estimates, a null distribution under the hypothesis of no difference between the sites was calculated by permutation testing with 10 000 permutations. The P-values represent the proportion of the permuted values that had pairwise statistics larger than the observed value (Michalakis & Excoffier 1996). Isolation by distance was examined by plotting pairwise θ/(1−θ) values against the log of the Euclidean geographical distances and was tested using the Mantel test with permutations as implemented in genetix.
Analysis of molecular variation (amova) was used to estimate variance components and to test the significance of partitioning of microsatellite variation within and among sites and regions. Observations are grouped into levels of a hierarchy; for example, individuals are grouped into populations, and populations into regions. The percentage of the overall variance that each level explains was then used to determine the significance of the groupings (Excoffier et al. 1992). amova was performed using genalex v.5 (Peakall & Smouse 2001). In this case variation was partitioned into two regions, to the northwest and southeast of the migratory divide, as well as into the sites within regions and individuals within sites.
Plumage analysis
The breeding plumage of male queleas is conspicuous and extremely variable from individual to individual. The facial mask around the beak can be black, brown or white. The colour of the crown, breast and belly can vary from light straw to deep pink, although characteristically in southern African Q. q. lathamii the belly is grey. Eight plumage characters were used to describe the observed variation in male plumage patterns. A total of 1397 queleas were assessed from 19 of the 23 sites (Table 1). Plumage characters were selected to include all the main areas of quelea plumage that show colour variation (Sinclair et al. 1993). The measure RMI (reduced mask index) represented the width of the black or white plumage mask around the head, which was scored: 0 (white-faced individual), 1 (black facial mask not extending above bill), 2 (thin black band above bill) and 3 (a thick black band of > 2 mm extending above the bill). This is similar to the mask index proposed by Ward (1966). Breast colour was scored from 1 (buff) to 4 (pink), and when this measure was buff, breast buff was scored from 1 (light) to 3 (dark). Belly grey recorded the extent of grey scaly feathers on the belly, from 1 (none) to 4 (full). Belly colour recorded the underlying colour of the feathers on the belly, from 0 (grey) through 1 (buff) to 3 (pink). Belly colour intensity was scored from 0 (pale) to 3 (dark). Crown colour and crown buff recorded the colour on the crown of each bird and were scored in the same way as the breast colour.
These plumage characters were used in a principal components analysis (PCA), carried out in minitab, to reduce complex multidimensional patterns of variation to a few principal components without losing a great deal of information (Afifi & Clark 1996). In order to examine any potential relationship between plumage variation and genetic variation, a Mantel permutation test was performed in genetix. Pairwise genetic distance was compared to the plumage distance between individuals. This was calculated as the straight-line distance between the location of individuals plotted on a graph of the first two principal components.
Results
Genetic differentiation
Observed (HO) and expected (HE) heterozygosity and values of FIS were estimated for all sites. Observed heterozygosity was generally high, ranging from 0.544 for MA/Phtr2 (site code/microsatellite locus) to 0.97 for TU/Esc4. Locus WBSW4 was the most heterozygous, with HO ranging between 0.80 and 0.96 (Table 2). FIS ranged from −0.061 for KR/Hru5–0.188 for NN/Mcyu4. Allele frequencies, FIS, HO and HE for all loci are given in Appendix I.
After sequential Bonferroni correction for multiple tests, three site-by-locus comparisons were significantly different from Hardy–Weinberg expectations at the 0.05 level. For locus Mcyu4, one site (LT) showed significant deviation from equilibrium, and two sites (LM and MA) were not in equilibrium for locus Phtr3. All the deviations were due to heterozygote deficiency. Exact tests for genotypic linkage disequlibrium showed two cases (Phtr2 and WBSW4 and Phtr3 and Hru5) of significant nonrandom associations between loci. As there were no consistent patterns of deviation from equilibrium by locus or site, all were included in further analyses.
Fisher's exact test showed that there was significant population differentiation across all sites (χ2 = 106.73, d.f. = 16, P < 0.001). However, the distribution of pairwise comparisons for θ was uniformly low, ranging from −0.026 for the BU–AF comparison to 0.008 for the JD–AF comparison. Most values were at or around zero with a many negative values. Negative values of θ indicate that there was more variance within the sites than there was between them and essentially indicates that there was no differentiation among sites. After correcting for multiple tests there were no significant differences between sites. In a smaller study on females (eight sites, n= 170) there were similarly low levels of genetic differentiation (range −0.036–0.008) (Dallimer et al. 2002). Hence we do not believe that concentrating on males in this study has biased the results in any way.
The Mantel test indicated that there was no significant isolation by distance (P = 0.488). The amova also revealed no significant difference between sites either side of the migratory divide (var = 0.000, P = 0.291; Table 3): 99% of the total variation was at the individual level within sites, with only 1% of the variation between sites within regions. None of the variation was between the two defined regions either side of the migratory divide.
| Source of variation | d.f. | Sum of squares | Components of variance | Percentage variation | P-value |
|---|---|---|---|---|---|
| Between regions | 1 | 5.737 | 0.000 Va | 0.0 | 0.291 |
| Among sites within regions | 16 | 87.571 | 0.023 Vb | 1.0 | 0.010 |
| Among individuals within sites | 1590 | 5444.254 | 3.424 Vc | 99.0 | 0.001 |
| Total | 1607 | 5537.562 | 3.447 |
Plumage
Three of the eight plumage characters, belly grey, belly colour and belly colour intensity, showed significant variation at the 0.05 level after sequential Bonferroni correction for multiple tests (Table 4). In a PCA the first two principal components contained 67% of the total variance. The sample means and standard errors for the first two principal components are given in Table 5, and illustrated in a scatter plot in Fig. 3.
| Character | H | d.f. | P |
|---|---|---|---|
| Belly grey | 63.64 | 18 | < 0.001 |
| Belly colour | 62.63 | 18 | < 0.001 |
| Belly colour intensity | 61.32 | 18 | < 0.001 |
| Breast colour | 19.33 | 18 | 0.372 |
| Breast buff | 14.22 | 18 | 0.714 |
| Crown colour | 19.16 | 18 | 0.382 |
| Crown buff | 21.01 | 18 | 0.279 |
| RMI | 9.60 | 18 | 0.944 |
| Site | n | PC1 | PC2 | |||
|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | |||
| Alwyn Farm | AF | 53 | −0.266 | 0.243 | 0.539 | 0.196 |
| Eden Farm | ED | 43 | −0.102 | 0.346 | 0.187 | 0.153 |
| Gumare | GU | 63 | 0.045 | 0.262 | 0.026 | 0.172 |
| Kroonstad | KR | 57 | −0.226 | 0.227 | −0.552 | 0.146 |
| Klawervallei | KW | 62 | −0.414 | 0.243 | 0.837 | 0.195 |
| Lichtenberg | LT | 70 | −0.366 | 0.227 | −0.379 | 0.119 |
| Maitengwane | MA | 79 | 0.035 | 0.216 | −0.215 | 0.130 |
| Nokoneng North | NN | 80 | 0.158 | 0.226 | 0.371 | 0.147 |
| Nokoneng South | NS | 59 | 0.657 | 0.242 | 0.394 | 0.185 |
| Riverside Farm | RF | 90 | 0.450 | 0.199 | 0.256 | 0.134 |
| Reata Ranch | RR | 98 | 0.117 | 0.204 | −0.062 | 0.110 |
| Shirville Farm | SH | 96 | −0.304 | 0.189 | −0.187 | 0.142 |
| Terminus | TE | 92 | 0.270 | 0.203 | −0.057 | 0.129 |
| Tuinplaas | TU | 55 | −0.381 | 0.230 | −0.354 | 0.176 |
| White Kopjes | WK | 33 | −0.104 | 0.305 | −0.411 | 0.192 |
| Senuko | XA | 88 | 0.019 | 0.201 | −0.088 | 0.152 |
| Bumi Hills | XB | 97 | −0.036 | 0.181 | 0.173 | 0.148 |
| Maitengwe Dam | XC | 90 | 0.075 | 0.200 | 0.029 | 0.137 |
| Malilangwe | XX | 92 | −0.027 | 0.180 | −0.439 | 0.144 |
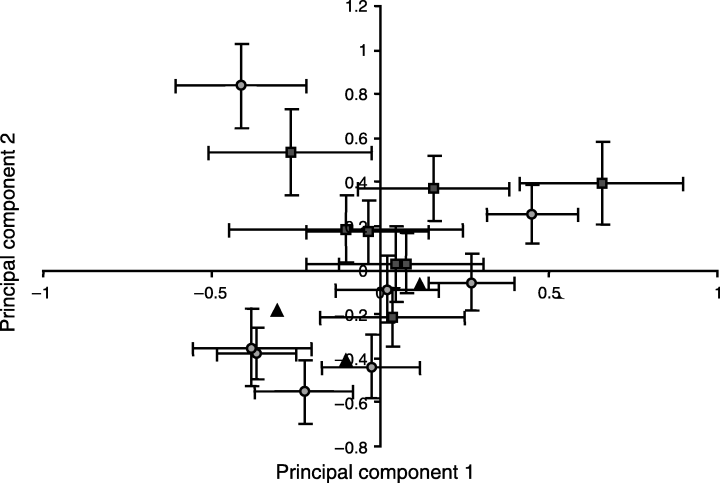
Scatter plot showing the first two principal components of plumage character variation between sample means. PC1 represents colour variation from buff (negative values) to pink (positive values). PC2 represents variation from not deeply coloured underside (negative) to coloured underside (positive). Samples from the northwest of the migratory divide are shown as squares, those from the southeast as circles. Other sites, from the Central region are triangles. The error bars represent standard errors of the sample means.
Principal component one (PC1) was determined mainly by a combination of four characters: breast colour, breast buff, crown colour and crown buff (Table 6). A positive PC1 indicated high values for the colour scores on the breast and crown, i.e. increasing pink colouration. A negative PC1 therefore indicated birds that were more buff, and as both crown buff and breast buff had high negative eigenvalues, where birds were scored as buff, they were also a deeper shade of buff than birds with a positive PC1. Hence PC1 can be viewed as an axis expressing the variation in colour around the facial mask from dark buff to pink.
| PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 | ||
|---|---|---|---|---|---|---|---|---|---|
| (a) | Eigenvalue | 3.584 | 1.771 | 1.014 | 0.682 | 0.428 | 0.244 | 0.177 | 0.101 |
| Proportion | 0.448 | 0.221 | 0.127 | 0.085 | 0.053 | 0.030 | 0.022 | 0.013 | |
| Cumulative | 0.448 | 0.669 | 0.796 | 0.881 | 0.935 | 0.965 | 0.987 | 1.000 | |
| Character | |||||||||
| (b) | Belly grey | −0.129 | −0.492 | 0.162 | 0.840 | 0.085 | 0.000 | −0.038 | −0.001 |
| Belly colour | 0.293 | 0.530 | 0.131 | 0.360 | −0.008 | −0.241 | 0.655 | 0.018 | |
| Belly colour intensity | 0.310 | 0.525 | 0.115 | 0.298 | 0.027 | 0.079 | −0.720 | −0.028 | |
| Breast colour | 0.477 | −0.168 | −0.020 | 0.006 | −0.207 | 0.563 | 0.112 | 0.610 | |
| Breast buff | −0.419 | 0.254 | 0.048 | 0.004 | 0.724 | 0.229 | 0.051 | 0.424 | |
| Crown colour | 0.459 | −0.168 | −0.003 | −0.077 | 0.539 | 0.352 | 0.124 | −0.570 | |
| Crown buff | −0.434 | 0.271 | 0.058 | 0.124 | −0.366 | 0.665 | 0.145 | −0.350 | |
| Reduced mask index | 0.003 | −0.084 | 0.968 | −0.233 | −0.033 | −0.015 | −0.006 | 0.012 | |
Principal component two (PC2) was mainly determined by variation in the colour of the belly (belly grey, belly colour and belly colour intensity) (Table 6). A positive PC2 indicated individuals with pinker, more deeply coloured bellies, while a negative PC2 indicated individuals with more grey on the belly and less colour. This axis can therefore be seen as expressing the variation in colouration on the underside of birds. The Mantel test showed that there was no relationship between plumage variation, as calculated above, and genetic variation (P = 0.8945).
There was no significant difference between the PC1 of samples from either side of the migratory divide (t-test: t = 0.9727, d.f. = 1168, P = 0.3309). However, there was a significant difference for PC2 (t-test: t = 3.2859, d.f. = 1168, P= 0.001), with individuals northwest of the divide having higher PC2 values, indicating pinker, more deeply coloured bellies. The scatter plot (Fig. 3) revealed no distinct clusters, and although there was a tendency for locations from the northwest to be found in the positive part of the plot, the site means were tightly clumped with a large overlap in their standard error bars in the centre of the plot. As PC2 represents only 22% of the of the total morphological variance and the overall plot of principal components shows a great deal of overlap between sites, we do not believe that this statistical difference represents a real biological phenomenon.
Discussion
Despite the recent experimental evidence that queleas may migrate in two different directions at the onset of the rains (Dallimer & Jones 2002), and earlier indications that flocks can remain cohesive over some months during migration (Jaeger et al. 1986), we found no evidence of significant genetic structure among red-billed queleas in southern Africa across the hypothesized migratory divide. Individuals with apparently similar genotypes occurred throughout the sampled range and it was impossible to identify any signal of genetic differentiation. There was also no evidence for geographical variation in plumage patterns across southern Africa; the main plumage polymorphisms were present at similar frequencies in each site studied. Although there was some evidence that queleas had more deeply coloured bellies to the northwest of the migratory divide, the overall evidence points to a lack of plumage differences between sites on either side of it.
There are two main reasons why we may not find differences across the migratory divide. First, the divide may not act to separate individuals into populations between which genetic and plumage differences can become established. Second, the molecular markers that we used were not appropriate for the task. We discuss each reason in turn.
We suggest that although a migratory divide at the start of the wet season in central southern Africa may act to separate queleas into two discrete geographical populations for their first breeding attempt of the season, birds from the two populations are likely to meet again later in the breeding season. This is because their subsequent breeding migrations bring them back towards the interior of the subcontinent, where they make their second and later breeding attempts (Fig. 1). Breeding colonies are then likely to contain individuals originating from many sources and from either side of the migratory divide, with the result that no genetic substructure can develop.
Birds in general show low amounts of genetic differentiation because of their high dispersal abilities (Crochet 1996). Nonetheless, the existence of population structure should not be ruled out simply because the species in question is wide-ranging and can migrate over long distances. Instead it is important to be aware of the limitations of using genetic techniques with species that could exhibit high levels of gene flow. In such species, where the signal relating to any population structure is going to be weak, sources of noise and error become more important and could bias the estimates of gene flow (Waples 1998).
Microsatellites may not be the best tool under such circumstances as they often show weak differentiation (Wennerberg et al. 1998; Petit & Mayer 1999; Nesje et al. 2000; Helbig et al. 2001). In our case, for most of the loci used the most common allele was the same in all sites tested. One of the main advantages of microsatellites, however, is that they show a range of polymorphisms from which appropriate makers can be chosen for the ecological problem in hand. Although we chose what a pilot study indicated to be suitable loci, the amount of variation revealed by all the loci may still have been too high and obscured any pattern that may have been present. Hence the isolation and use of microsatellites with fewer repeats (< 10) may have been more appropriate.
Despite these difficulties, however, our results are in accordance with other direct observational studies, which also point to a lack of structure in southern Africa (Jaeger et al. 1986; Bruggers 1989; Johns et al. 1989; Mundy & Herremans 1997; Oschadleus 2000). We conclude that while there is evidence that the migratory divide exists, it does not result in genetic and plumage variation in the populations to either side of it.
This result has some practical importance. The red-billed quelea is one of Africa's most destructive vertebrate pests, accounting for the loss of up to 5% of small-grain cereal output (Elliott & Lenton 1989). Many countries maintain quelea control programmes (e.g. Geertsema 1999) that may be managed more effectively if better information were available relating to population structure and movement of quelea in the region. Our results confirm that queleas in southern Africa should continue to be managed as a single population. Furthermore, they imply that the recommended monitoring technique of using male plumage morph frequencies to track quelea movements (Allan 1996) is uninformative in southern Africa and there is little benefit in continuing its use there.
Acknowledgements
We would like to thank all those who helped with sample collection; in Zimbabwe: Peter Mundy, Ngoni Cheweshe, Jim Dale, Bruce Davidson and Douglas Kabale; in South Africa: Craig Whittington-Jones, Luka Geertsema, Jonas Örnborg, Nicki and Paul Vet and Steven Piper; in Botswana: T.S. Moruti and Mr Galekgowe; and in Namibia: Berthold Wohlleber and Rob Simmons. Stuart Piertney provided useful comments on an earlier draft of the manuscript. This project was supported by a NERC CASE studentship to MD and was partly funded by the Crop Protection Programme (Project R6823) of the United Kingdom Department for International Development (DFID). The views expressed are not necessarily those of DFID.
Supplementary material
The following material is availiable from http://www.blackwellpublishing.com/products/journals/suppmat/MEC/MEC1733/MEC1733sm.htm
Appendix I
Table A1. Microsatellite allele frequency distribution, observed and expected heterozygosities and FIS population and locus.
References
This study formed the core of Martin Dallimer's PhD, and was carried out in Josephine Pemberton's molecular ecology laboratory. It formed part of a project led by Bob Cheke in collaboration with Peter Jones to forecast quelea movements in relation to rainfall patterns across southern Africa.




