Microsatellite variation in natural Drosophila melanogaster populations from New South Wales (Australia) and Tasmania
Abstract
Microsatellite variation was studied at 48 microsatellite loci in 10 Drosophila melanogaster populations to investigate the population structure on the Australian east coast. Low, but statistically significant population differentiation was observed among most populations. The populations on the Australian mainland did not show evidence for isolation by distance. We conclude that the population structure of D. melanogaster on the Australian mainland is probably the result of a shared history (recent colonization). The observed differences between local D. melanogaster populations probably reflect variation in effective population sizes rather than patterns of gene flow. Two populations from Tasmania were more differentiated from the Australian mainland than a population from Israel, raising the question whether they are derived from the Australian mainland or colonized from a different source population.
Introduction
Drosophila melanogaster is one of the most prominent genetic model organisms. With the availability of the complete genomic sequence, D. melanogaster research is about to change its emphasis. The isolation of novel genes will no longer be required, but the characterization of gene function will be one of the primary research targets. While the characterization of single genes, expression profiling and proteomics will be the major avenues, the exploitation of natural variation could also be an important contribution to functional genomics (Alonso-Blanco & Koornneef 2000).
To take full advantage of the naturally occurring variation, a solid understanding of the population structure and history is required. Despite the full body of literature available for D. melanogaster, the structure of natural populations has not been extensively studied. It is widely accepted that D. melanogaster has its origin in tropical Africa and colonized the Eurasian continent approximately 10 000–15 000 years ago, after the end of the last glaciation (David & Capy 1988). Other continents were subsequently colonized. The earliest reports of D. melanogaster from the Australian continent date approximately 100 years (Boussy et al. 1998) back.
Mark and recapture experiments suggest that D. melanogaster has a high capacity to migrate (Coyne et al. 1982). Hence, it has been assumed that D. melanogaster is an almost panmictic species. Nevertheless, early allozyme studies indicated some population differentiation within and between continents (Singh et al. 1982; Singh & Rhomberg 1987). Because allozymes could be the target of selection, it was not clear whether a population differentiation as indicated by allozymes argues against the hypothesis of a panmictic species. One particularly illustrative work examined this question further. On the east coast of northern America a cline has been described for the fast/slow Adh polymorphism (Berry & Kreitman 1993). The authors used a restriction fragment length polymorphism (RFLP) analysis of the Adh gene in populations along the east coast. The single nucleotide polymorphism (SNP), which causes the fast/slow amino acid replacement, fitted the expectation based on allozyme studies. The silent polymorphisms, however, did not show a clinal variation. The conclusion from this pattern was that high migration rates prevented a differentiation of the populations, but selection maintained the allozyme polymorphism (Berry & Kreitman 1993). Hence, this interpretation was consistent with the general definition of a cline, which is assumed to be maintained by natural selection despite high migration rates (Endler 1977). Since then, an increasing number of studies have found evidence for population substructuring in D. melanogaster, thus challenging the idea of a panmictic species (Begun & Aquadro 1993, 1995; Schlötterer et al. 1997; Aguadé 1999).
The observed clinal variation on the Australian east coast (Oakeshott et al. 1982; Anderson et al. 1987; Boussy & Kidwell 1987; James et al. 1997; Gilchrist & Partridge 1999; McColl & McKechnie 1999), in combination with the use of a large number of neutrally evolving markers such as microsatellites, provides a good opportunity to differentiate between potentially operative mechanisms responsible for observing (maintaining) population structure. Here we present a data set based on 48 microsatellite loci screened in nine Australian D. melanogaster populations.
Materials and methods
Isofemale lines
In 1997, I. A. Boussy and R. Woodruff collected Drosophila melanogaster flies along the east coast of Australia (Fig. 1). Since then, they have been maintained as isofemale lines. The lines used in this study were collected in New South Wales: in Laurieton (31.648° SL, 28 lines), Forster (32.179° SL, 34 lines), Wootton (32.263° SL, 34 lines), Malua Bay (35.792° SL, 18 lines), Moruya (35.910° SL, 32 lines), Tuross Head (36.060° SL, 12 lines), Nageela Orchard (36.160° SL, 31 lines); and in Tasmania: Trial Bay Orchards (43.138° SL, 24 lines) and Cygnet (43.161° SL, 29 lines). The Israeli flies were the F1 generation from single wild females caught in 1997 at the Nahal Oren Canyon, Israel, by A. Kamal. Genomic DNA was extracted from single individuals using a high salt extraction method (Miller et al. 1988).
Geographical map from eastern Australia with the locations and names of the sampling sites along the east coast and Tasmania.
Microsatellite amplification
The microsatellite loci used in this study were either isolated from P1 clones or retrieved from the Drosophila genome project web pages. The loci used in this study are listed in Table 1, and further information is available from http://i122server.vu-wien.ac.at. The microsatellite loci were amplified in 10 µL reactions (1.5 mm MgCl2, 2 µm each primer and 0.5 U Taq polymerase) following standard protocols (Schlötterer 1998). Five minutes of initial denaturation at 94 °C was followed by 30 cycles of 50 s at 94 °C, 50 s at 40–55 °C (depending on the primer combination) and 50 s at 72 °C. We used a final extension at 72 °C for 45 min to ensure quantitative terminal transferase activity of the Taq polymerase. Polymerase chain reaction (PCR) products were separated on a 7% denaturing polyacrylamide gel (32% formamide, 5.6 m urea). The PCR products were sized by running a sizing ladder next to the amplified microsatellites (Schlötterer & Zangerl 1999).
| Locus | Chromosome | Number of alleles | V̄ | H̄ | Locus | Chromosome | Number of alleles | V̄ | H̄ |
|---|---|---|---|---|---|---|---|---|---|
| 3641.2 | x | 15 | 2.196 | 0.605 | Dm1639-CA | 2 | 10 | 4.263 | 0.566 |
| DS06335b | x | 6 | 0.274 | 0.077 | Dm1639-TA | 2 | 7 | 0.709 | 0.589 |
| Dmtena | x | 8 | 2.360 | 0.706 | Dm1639-TC | 2 | 9 | 1.771 | 0.633 |
| DS00146 | x | 3 | 0.224 | 0.421 | Dm2337 | 2 | 11 | 2.612 | 0.706 |
| DS00265a | x | 6 | 1.640 | 0.464 | Dmmp20 | 2 | 6 | 2.120 | 0.393 |
| DS00265b | x | 4 | 1.128 | 0.373 | Dpt-GT | 2 | 9 | 1.564 | 0.544 |
| DS00314 | x | 2 | 0.008 | 0.016 | Dpt-TA | 2 | 12 | 2.853 | 0.743 |
| DS00589 | x | 4 | 8.277 | 0.542 | Drogpad | 2 | 13 | 1.864 | 0.489 |
| DS01551 | x | 6 | 0.203 | 0.439 | Drogpdha | 2 | 6 | 0.202 | 0.475 |
| DS09020 | x | 9 | 0.902 | 0.501 | Droninac | 2 | 4 | 0.410 | 0.522 |
| DS09021 | x | 17 | 27.485 | 0.688 | DS001340 | 2 | 12 | 1.038 | 0.552 |
| Z31849 | 2 | 5 | 2.119 | 0.454 | DS00144 | 2 | 3 | 0.027 | 0.132 |
| 6744 | 2 | 5 | 0.260 | 0.428 | DS09065/1 | 2 | 7 | 5.574 | 0.645 |
| DS08011 | 2 | 9 | 8.434 | 0.758 | DS09065/2 | 2 | 7 | 8.517 | 0.734 |
| Adh-TC | 2 | 13 | 6.220 | 0.716 | Eno-CA | 2 | 8 | 2.838 | 0.623 |
| Cact-TA | 2 | 5 | 1.541 | 0.526 | Eno-TA | 2 | 8 | 0.993 | 0.674 |
| Cad-CA | 2 | 12 | 1.216 | 0.566 | Ft-CA | 2 | 3 | 6.175 | 0.555 |
| Cad-GA | 2 | 7 | 0.770 | 0.329 | Ft-TA | 2 | 4 | 0.112 | 0.223 |
| Cad-TA | 2 | 7 | 2.776 | 0.685 | G410 | 2 | 32 | 19.085 | 0.805 |
| Cd36 | 2 | 3 | 0.340 | 0.063 | G411 | 2 | 3 | 0.158 | 0.279 |
| Dm00600CA | 2 | 7 | 0.807 | 0.487 | Pkc53E-CA | 2 | 8 | 1.273 | 0.635 |
| Dm00600TC | 2 | 9 | 2.270 | 0.414 | Pkc53E-GA | 2 | 9 | 1.182 | 0.600 |
| Dm0332-CA | 2 | 4 | 2.095 | 0.172 | Z32225 | 2 | 5 | 0.360 | 0.179 |
| Dm0600-TA | 2 | 10 | 0.294 | 0.491 | Z50409 | 2 | 7 | 4.803 | 0.682 |
- V̄, variance in repeat number; H̄, average heterozygosity.
Data analysis
General measures of genetic variation, such as heterozygosity and variance in allele sizes, were calculated using the microsat 1.5 software package (Minch et al. 1995) and corrected for small sample sizes (Nei & Roychoudhury 1974). To estimate gene flow and population differentiation, pairwise Θ values, as an unbiased estimate of FST (Weir & Cockerham 1984), were generated in the fstat software package (Goudet 1995). In the following we will refer to Θ values as FST values. For the isofemale lines, one allele was randomly selected to account for genetic drift during the propagation of the lines. In the case where a diploid data set was required, all alleles were doubled to create a diploid data set. The significance of pairwise FST values was tested by permuting genotypes among populations (10 000 times), as this method does not rely on Hardy–Weinberg assumptions (Goudet et al. 1996). The significance of pairwise FST values was corrected for multiple testing using a sequential Bonferroni technique (Sokal & Rohlf 1995).
To test for a significant correlation between genetic and geographical distances we compared an FST/(1 − FST) matrix with a geographical distance matrix (ln km) (Rousset 1997) using the Mantel test (10 000 permutations) in the genepop program (Raymond & Rousset 1995). Geographical distances (in km) between sampling sites were obtained using global positioning system (GPS) coordinates on a distance calculator (http://www.nau.edu/~cvm/latlongdist.html).
Genetic distances were calculated on the proportion of shared alleles as implemented in microsat. The obtained distance matrix was converted into a dendrogram using the upgma algorithm provided with the phylip software package (Felsenstein 1991) and graphically displayed with treeview (Page 1996).
Principal component analysis on the variance–covariance matrix of allele sizes was performed in spss. Individuals were a priori grouped into both populations and regions.
The software bottleneck (Cornuet & Luikart 1996) was used to infer population bottlenecks.
Results
Forty-eight microsatellite loci were analysed in 10 natural Drosophila melanogaster populations, one originating from Israel and nine Australian populations. Large differences in variability were observed between the loci analysed. The number of alleles ranged from two to 32, with an average of 7.9 (Table 1). Similarly, other measurements of variability differed among the analysed loci. Gene diversities averaged 0.5, ranging from 0.016 to 0.81. The largest difference among loci was observed for the variance in repeat number, a widely used diversity measurement for microsatellites (Goldstein & Schlötterer 1999). The variance in repeat number ranged from 0.008 to 27.5 and averaged 3.0.
Among populations, average gene diversities were similar, ranging from 0.45 to 0.52 (Table 2). Variances in repeat number (averaged over loci) varied between 2.3 and 3.3 (Table 2). Irrespective of the measurement used, two populations from Tasmania showed the lowest levels of variability. The population from Israel had levels of variation very similar to the Australian mainland populations.
| Population | V̄ | H̄ | n |
|---|---|---|---|
| Laurieton | 3.045 | 0.517 | 28 |
| Forster | 3.336 | 0.523 | 34 |
| Wootton | 2.978 | 0.506 | 35 |
| Malua Bay | 3.258 | 0.492 | 18 |
| Moruya | 2.589 | 0.511 | 32 |
| Tuross Head | 3.867 | 0.492 | 12 |
| Nageela Orchard | 2.905 | 0.506 | 31 |
| Cygnet | 2.290 | 0.455 | 29 |
| Trial Bay Orchards | 2.426 | 0.451 | 24 |
| Israel | 3.193 | 0.520 | 58 |
- V̄, variance in repeat number; H̄, average heterozygosity; n, number of isofemale lines.
Population structure
Consistent with previous studies, we detected low levels of population substructuring (FST = 0.039). Nevertheless, FST values were significantly different from zero, irrespective of whether all Australian populations were included jointly (FST = 0.035) or only Australian mainland populations were analysed (FST = 0.015). To determine the influence of geographical distance on population differentiation, we employed a specific sampling regime. Multiple populations were included from three geographical regions separated by at least 432 km. Within each geographical region, the analysed populations were located no more than 65.1 km apart from each other. The combined analysis of all populations from the Australian mainland and Tasmania suggested a significant effect of geographical distance on genetic differentiation (Mantel test, P = 0.0095). The comparison of FST values with and without Tasmanian populations, however, indicated that most of the genetic differentiation was contributed by the Tasmanian flies. Pairwise FST values between Tasmanian and Australian mainland populations ranged from 0.053 to 0.099, while comparisons within the Australian mainland were almost an order of magnitude lower (Table 3). The significance of the genetic differentiation between Tasmania and mainland Australia can be put into perspective using the population from Israel. Average FST values between Tasmania and Australian mainland populations were higher (FST = 0.071 ± 0.012) than between Israel and mainland Australia (FST = 0.046 ± 0.005).
| North | Central | South | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Laurieton | Forster | Wootton | Malua Bay | Moruya | Tuross Head | Nageela Orchard | Cygnet | Trial Bay Orchards | |
| Forster | 0.017* | ||||||||
| Wootton | 0.027* | 0.009NS | |||||||
| Malua Bay | 0.031* | 0.032** | 0.025* | ||||||
| Moruya | 0.020* | 0.021* | 0.027** | 0.026* | |||||
| Tuross Head | 0.021NS | 0.003NS | 0.020NS | 0.035* | 0.013NS | ||||
| Nageela Orchard | 0.026** | 0.009NS | 0.016* | 0.031* | 0.019* | 0.017NS | |||
| Cygnet | 0.077** | 0.061** | 0.073** | 0.077** | 0.056** | 0.070** | 0.053** | ||
| Trial Bay Orchards | 0.082** | 0.085** | 0.082** | 0.099** | 0.065** | 0.088** | 0.067** | 0.033* | |
| Israel | 0.048** | 0.040** | 0.045** | 0.057** | 0.044** | 0.044** | 0.050** | 0.083** | 0.094** |
- NS, not significant; *P = 0.05; **P = 0.01.
Given the large influence of the Tasmanian flies, we repeated the analysis for the association between geographical distance and genetic differentiation for Australian mainland populations only. Using seven populations separated by up to 565 km, we did not detect a significant association between genetic and geographical distance. Similarly, principal component analysis also failed to reveal geographical differentiation within the Australian mainland (data not shown). Inspection of pairwise genetic distances (Table 3) indicates why. Pairs of populations, such as Laurieton and Wootton, which are located only 86.4 km apart (Table 4), show significant FST values, while others (Nageela Orchard and Forster), separated by about 500 km, are not significantly different from each other.
| Laurieton | Forster | Wootton | Malua Bay | Moruya | Tuross Head | Nageela Orchard | Cygnet | |
|---|---|---|---|---|---|---|---|---|
| Forster | 65.1 | |||||||
| Wooton | 86.4 | 24.2 | ||||||
| Malua Bay | 517.8 | 452.7 | 432.0 | |||||
| Moruya | 537.3 | 472.3 | 451.4 | 20.1 | ||||
| Tuross Head | 17.3 | 484.8 | 464.3 | 32.8 | 17.3 | |||
| Nageela Orchard | 564.8 | 499.7 | 479.4 | 49.1 | 15.5 | 16.7 | ||
| Cygnet | 1361.6 | 1296.6 | 1277.5 | 849.0 | 831.0 | 816.3 | 800.4 | |
| Trial Bay Orchards | 1368.7 | 1303.8 | 1284.9 | 857.5 | 839.5 | 851.9 | 808.7 | 14.7 |
To verify the obtained pattern, we used genetic distance as an additional measurement of differentiation between populations. Using the proportion of shared alleles as a measure of the genetic similarity of D. melanogaster populations we obtained similar results as with the FST analysis. Figure 2 indicates that two clades are supported by high bootstrap values. The two Tasmanian populations group together with 100% bootstrap support and the populations from Wootton, Forster and Nageela Orchard also group together (78%). Both groupings were also detected using neighbour joining as the clustering algorithm (not shown). Interestingly, while the populations from Wootton and Forster were sampled in close proximity, the other population was collected at Nageela Orchard approximately 500 km away. Hence, consistent with the FST analysis, the proportion of shared alleles also indicates that the differentiation of D. melanogaster populations on the Australian mainland does not follow a simple isolation-by-distance model.
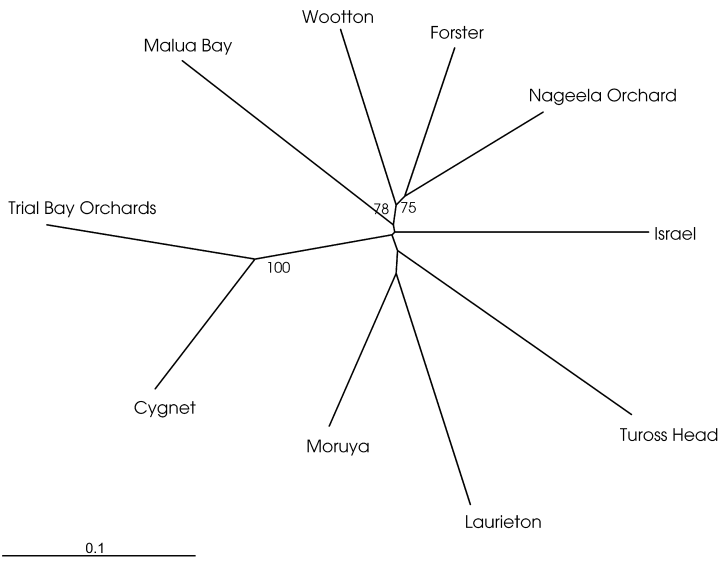
Unrooted upgma phylogram of populations based on the proportion of shared alleles. The numbers written next to the nodes indicate bootstrap values from 100 replications (only bootstrap values above 70 are displayed).
Bottleneck
Recently, the difference between observed gene diversity and the gene diversity expected on the number of observed alleles has been suggested as a test for bottlenecks/population expansion (Cornuet & Luikart 1996). Applying this test to our data yielded inconclusive results (Table 5). Depending on the mutation model and the test statistic, different results were obtained. However, significant test statistics were not confined to the Tasmanian populations, but also detected in the Australian mainland and Israeli populations. Similar results were obtained when we divided the analysed populations into three groups: Australian mainland, Tasmania and Israel (data not shown). Therefore, we conclude that this test statistic does not support the hypothesis of a more extensive bottleneck in the Tasmanian populations than in the other populations analysed.
| Population | Mutation model | Observed heterozygosity | Sign test | Standardized difference | Wilcoxon |
|---|---|---|---|---|---|
| Cygnet | IAM* | H E excess | 0.00217 | 0.00000 | 0.00000 |
| TPM† | H E excess | 0.05072 | 0.00088 | 0.00051 | |
| SMM‡ | H E excess | 0.49800 | 0.40481 | 0.48332 | |
| Trial Bay Orchards | IAM | H E excess | 0.23046 | 0.02714 | 0.02839 |
| TPM | H E excess | 0.45168 | 0.30110 | 0.29036 | |
| SMM | H E deficiency | 0.00733 | 0.00130 | 0.01479 | |
| Laurieton | IAM | H E excess | 0.00016 | 0.00005 | 0.00000 |
| TPM | H E excess | 0.03912 | 0.01708 | 0.00259 | |
| SMM | H E deficiency | 0.10992 | 0.01372 | 0.13653 | |
| Forster | IAM | H E excess | 0.00153 | 0.00030 | 0.00004 |
| TPM | H E excess | 0.10828 | 0.06082 | 0.04791 | |
| SMM | H E deficiency | 0.02218 | 0.00029 | 0.02388 | |
| Wootton | IAM | H E excess | 0.00481 | 0.00087 | 0.00055 |
| TPM | H E excess | 0.47460 | 0.10627 | 0.10962 | |
| SMM | H E deficiency | 0.00716 | 0.00002 | 0.01279 | |
| Malua Bay | IAM | H E excess | 0.00021 | 0.00009 | 0.00001 |
| TPM | H E excess | 0.00944 | 0.00867 | 0.00187 | |
| SMM | H E excess | 0.23566 | 0.19973 | 0.52620 | |
| Moruya | IAM | H E excess | 0.00173 | 0.00060 | 0.00001 |
| TPM | H E excess | 0.41328 | 0.10718 | 0.04117 | |
| SMM | H E deficiency | 0.00002 | 0.00006 | 0.00135 | |
| Tuross Head | IAM | H E excess | 0.02535 | 0.00166 | 0.00007 |
| TPM | H E excess | 0.07799 | 0.03879 | 0.01113 | |
| SMM | H E excess | 0.26043 | 0.22602 | 0.68072 | |
| Nageela Orchard | IAM | H E excess | 0.02535 | 0.00039 | 0.00033 |
| TPM | H E excess | 0.42830 | 0.04203 | 0.03717 | |
| SMM | H E deficiency | 0.05077 | 0.00514 | 0.03908 | |
| Israel | IAM | H E excess | 0.16021 | 0.06687 | 0.01518 |
| TPM | H E deficiency | 0.01695 | 0.04159 | 0.16021 | |
| SMM | H E deficiency | 0.00000 | 0.00000 | 0.00000 |
- * IAM, infinite allele model;
- † TPM, two phase model (variance = 5.00, P = 20%);
- ‡ ‡SMM, stepwise mutation model.
Discussion
An implicit assumption of studies reporting clinal trait variation is that the pattern seen is being maintained despite gene flow, and thus serves as evidence for natural selection (Endler 1977). Because clinal variation on the east coast of Australia (Oakeshott et al. 1982; Anderson et al. 1987; Boussy & Kidwell 1987; James et al. 1997; Gilchrist & Partridge 1999; McColl & McKechnie 1999) is among the best studied in Drosophila melanogaster, we investigated the population structure along this cline. With the availability of microsatellites, we were, for the first time, able to use a polymorphic marker, which is probably evolving neutrally (Schlötterer 2000).
Like other studies, we also observed low levels of genetic differentiation among the Australian populations. Traditionally, these low levels of variability were attributed to the high dispersal capacities of D. melanogaster (Coyne et al. 1982). Hence, migration is assumed to prevent genetic differentiation at neutral markers. Previously it was shown that restricted migration results in a positive correlation between geographical and genetic distance (Slatkin 1993). To evaluate the impact of migration on the population structure of D. melanogaster, we designed a sampling regime that includes populations collected in close geographical proximity, as well as more distantly located populations. If migration was an important force shaping the population structure of D. melanogaster, one would expect either isolation by distance or no population differentiation at all. Our analysis on the Australian mainland, however, detected low, but statistically significant, population differentiation. Furthermore, the analysed populations showed no isolation by distance. Based on these results we conclude that simple migration models may not be sufficient to explain the partition of genetic variation on the Australian mainland.
Given that the high differentiation of adjacent populations combined with no significant differentiation between distantly located populations is not consistent with a simple migration model, an alternative explanation is required. While it is possible to construct more complex migration models, it is more parsimonious to consider a shared history of the populations as an explanation for the observed pattern of variability. Slatkin (1993) recently noted that the absence of isolation by distance could be indicative of a recent colonization event. A possible scenario could be that a relatively large founder population colonized Australia. Assuming that this population rapidly spread over Australia, a very similar allele distribution is expected over the entire continent. Subsequently, in the absence (or low levels) of migration, the populations gradually diverged from each other by genetic drift. Under this scenario, the magnitude of genetic differentiation between populations is therefore a function of their effective population size rather than geographical distance.
Tasmanian populations are differentiated from Australian mainland populations
A very interesting pattern emerged when the Australian mainland populations were compared with the two Tasmanian populations. Pairwise comparisons indicated that populations from the Australian mainland were less differentiated from each other than from those collected in Tasmania. A control population from Israel was not more differentiated from Australian mainland populations than the Tasmanian populations.
In principle, two alternative explanations for the high genetic differentiation between the Australian mainland and Tasmania could be given. Either the flies from Tasmania were colonized from the Australian mainland, but have gone through a major bottleneck, or they were colonized from a different geographical region. Some support for the bottleneck hypothesis comes from the lower genetic variability of the Tasmanian populations. All measurements of genetic variability were consistently lower in the Tasmanian populations. Recently, several test statistics were developed to infer population history from microsatellite data (Cornuet & Luikart 1996; Kimmel et al. 1998; Beaumont 1999; Reich et al. 1999). Unfortunately, the suspected population history of the Tasmanian populations is almost impossible to test. If the Tasmanian flies were derived from a moderate number of individuals from the Australian mainland population, which has also been recently founded, the population scenario is more complex than usually assumed for tests for population bottlenecks. Nevertheless, we applied test statistics comparing heterozygosity to the expected heterozygosity (based on the number of alleles) to our data set. The results were ambiguous. Depending on the mutation model assumed and the test statistic used, not only the Tasmanian populations, but also the Australian mainland and Israeli populations, had a statistically significant excess of heterozygosity for at least one mutation model/test statistic combination. When a Bonferroni correction was applied, the pooled Tasmanian populations had the same number of significant tests as the pooled Australian mainland population (data not shown).
Additional information about the history of populations could be gleaned from the genetic distance measurements. The phylogenetic tree based on the proportion of shared alleles clearly indicates that the two Tasmanian populations are more diverged from the Australian mainland populations than the population from Israel. Provided that the proportion of shared alleles has been successfully used for the phylogenetic reconstruction of closely related Drosophila species (Harr et al. 1998), we could exclude that the result is an artefact of the inclusion of too diverged populations. More intricate is the influence of a bottleneck on genetic distances. Bottlenecks are known to increase genetic distance values, which results in longer branch lengths (Chakraborty & Nei 1977). Figure 2 clearly indicates that the Tasmanian populations have the longest branch lengths, potentially suggesting a population bottleneck. This effect can be seen irrespective of the clustering algorithm used (not shown). Nevertheless, this observation does not preclude that the Tasmanian populations were colonized from a different source population than the Australian mainland (as this would also result in longer branch lengths). Further investigations are required to resolve the history of the Tasmanian populations. Recently, it has been shown that long microsatellites with a mutation rate similar to those observed for mammalian microsatellites can be found in D. melanogaster (Harr & Schlötterer 2000). Using these highly polymorphic loci may provide more insight into the population history of the Tasmanian flies.
Clines
Traits that vary along an environmental cline have long captivated evolutionary biologists, as such scenarios potentially provide an opportunity to study the adaptive response of populations to a selection gradient (Endler 1977). Latitudinal clines in various traits of D. melanogaster have been discovered, and these clines remain the subject of intense studies aimed at evaluating their genetic basis (Berry & Kreitman 1993; James et al. 1997; Gilchrist & Partridge 1999).
On the east coast of Australia, latitudinal clines are well described for phenotypic traits such as body size, cell number, cell area (James et al. 1997) and wing size (Gilchrist & Partridge 1999). Similar clinal variation was detected on a molecular basis. First, allozyme (Oakeshott et al. 1982) and inversion polymorphisms (Anderson et al. 1987) were described, but more recently sequence variation was detected at the heat shock hsr-omega (McColl & McKechnie 1999) gene. Boussy & Kidwell (1987) also reported a latitudinal cline in P-element-associated characteristics.
The fact that the pattern of clinal variation could be detected on different continents strongly suggests that selection rather than historical patterns shapes these clines. Hence, neutral markers should not exhibit clinal variation. To discriminate between a separate introduction of D. melanogaster from North America and Europe and selection, a mitochondrial DNA polymorphism was used for Australian D. melanogaster populations (Boussy et al. 1998). The authors showed that no clinal pattern could be detected and, therefore, the hypothesis of massive introduction from two continents could be ruled out (Boussy et al. 1998). In our data set, the distribution of microsatellite polymorphisms on the Australian mainland does not show clinal variation and the populations are only slightly diverged. Hence, the microsatellite data on the Australian mainland are consistent with the hypothesis that selection shapes the clines on the Australian east cost. Including the populations from Tasmania significantly complicates the situation. Provided that Tasmanian populations are well differentiated from Australian mainland populations, the question arises whether they are derived from the same ancestral population. As discussed above, the microsatellite data are ambiguous and do not allow a firm conclusion. On the other hand, selected variation showing a clinal pattern is also not conclusive, given that parallel clines were observed in different continents (Oakeshott et al. 1982). Hence, selection may have generated similar phenotypes in southern Australia and Tasmania, even if the populations originated from different colonization events. Interestingly, an a posteriori contrast analysis indicated that flies from Tasmania had larger wings with more cells than those from the Australian mainland (James et al. 1997). Furthermore, two Tasmanian populations showed very little genetic differentiation for wing size (Gilchrist & Partridge 1999).
One important aspect that emerges from our microsatellite analysis is that the clinal variation observed on the Australian mainland might not match a cline in its classical sense. In Australia, gene flow among populations seems to be low. Therefore, we suggest that most of the genetic variation seen along the Australian east coast may have already been present in the founder population(s). Differential selection could have resulted in the distribution of selected traits as it is currently observed. We would like to point out that this scenario could also apply to the North American east coast, where a clinal variation for Adh was recently described (Berry & Kreitman 1993). It is quite probable that sequence variability was too low in the studied sample to detect population differentiation. Hence, it would be highly informative to use a highly polymorphic marker, such as microsatellites, or a large set of SNP markers, to determine population structure at other natural clines.
Acknowledgements
We are grateful to I. A. Boussy, R. Woodruff and A. Kamal for providing flies. Special thanks to the members of the C.S. laboratory and in particular to S. Weiss for helpful comments. We appreciate discussions with M. Beaumont about microsatellite variation and bottlenecks. C.S. is supported through FWF grants.
References
This study is part of a large project in the laboratory of Christian Schlötterer, which aims to identify adaptive mutations in natural populations. Martin Agis has focused his PhD on D. melanogaster populations from Australia and Tasmania.




