Mode of reproduction and amplified fragment length polymorphism variation in purple needlegrass (Nassella pulchra): utilization of natural germplasm sources
Abstract
A dominant plant of the California grasslands, purple needlegrass [Nassella pulchra (Hitchc.) Barkworth] is an important revegetation species in its native range. The amplified fragment length polymorphism (AFLP) method was used to elucidate mode of reproduction and nucleotide variation among 11 natural populations and three selected natural germplasm releases of N. pulchra. A total of 12 co-dominant AFLPs, informative within eight populations, failed to reveal any heterozygous individuals, indicating very high selfing rates (S̄H = 1). Estimates of nucleotide diversity within populations ranged from 0 to 0.00069 (0.00035 average), whereas the total nucleotide divergence among populations ranged from 0.00107 to 0.00382 (0.00247 average). Measures of population differentiation (GS) in terms of Shannon–Weaver diversity values and estimated nucleotide substitutions were 0.90 and 0.86, respectively. Although some of the sample populations contained a mixture of true breeding genotypes, most populations could be distinguished unambiguously. Moreover, geographical distance between the natural source populations was significantly correlated with genetic distance (r = 0.60) among the corresponding sample populations. Results indicate that inbreeding, combined with founder effects and/or selection, has contributed to the differentiation of N. pulchra populations. Foundation seed populations of the selected natural germplasm releases were genetically well defined and most similar to natural seed collected near the corresponding source populations. Thus, these commercial germplasm sources will be made practically available and useful for conservation plantings within the intended areas of utilization.
Introduction
Grasses are the most commonly used plant materials for primary soil stabilization and weed exclusion in many large- and small-scale revegetation efforts. Grasses used for revegetation have profound impacts evident by the ecological and agricultural importance of introduced species, such as crested wheatgrass (Agropyron spp.), that were successfully used over the past century to stabilize soils and productivity on highly degraded rangelands and abandoned croplands of western North America. The release of commercial germplasm sources of native grass species, which will also be used in large-scale revegetation efforts, may have similar ecological and agricultural significance. Ecologists are concerned that the agronomic approach typically used for the production of these plant materials has been to select or breed cultivars with specific characteristics, which reduces genetic diversity (Roundy et al. 1997; Lesica & Allendorf 1999; Roundy 1999). Thus, many commercial sources of native plant materials may lack the genetic diversity needed to maintain adaptation in a natural environment. Moreover, nonlocal seed sources may not have adapted genomes and use of these plant materials may result in genetic pollution of natural populations (Roundy et al. 1997; Lesica & Allendorf 1999; Roundy 1999). As the demand for native grass seed has grown, so has the demand for source-identified germplasm thought to represent naturally diverse and/or well-adapted ecotypes suitable for different sites or niches. Yet one of the most serious impediments to the implementation of native plant materials, particularly in many large-scale applications, has been seed availability (Richards et al. 1998). Although commercial growers can produce large quantities of high-quality grass seed, the feasibility of producing readily available, source-identified seed for many different sites is another important consideration. Locally collected seed or propagates from natural populations may be the best approach for many plant species. However, it is inherently difficult to anticipate the amount of grass seed needed in many large-scale fire rehabilitation efforts, much less collect or increase this seed from the locally native populations.
Believed to have been a dominant plant of pristine California grasslands, Nassella pulchra (Hitchc.) Barkworth is still the most common native bunchgrass in northern California and is also widespread in southern California (Bartolome 1981; Bartolome & Gemmill 1981; Heady et al. 1991). Closely related species, N. lepida (Hitchc.) Barkworth and N. cernua (Stebbins & Love) Barkworth, share similar distributions (primarily within California) and occasionally hybridize with N. pulchra (Love 1954). Evidence suggests that N. pulchra (2n = 64) contains a full complement of N. lepida (2n = 34) chromosomes, however, N. cernua (2n = 70) contains only 14 of the 17 chromosome pairs in N. lepida (Love 1954). Although cytogenetic relationships are not fully understood, these species behave as normal diploids and are thought to be allopolyploid (Stebbins & Love 1941; Love 1954). Like many Stipeae species, N. pulchra produces copious quantities of large viable seeds with sharp calli and twisting awns. These traits enable N. pulchra to rapidly colonize or increase abundance on many disturbed sites (Bartolome 1981; Bartolome & Gemmill 1981). These characteristics also make N. pulchra an important restoration tool within its native California range (Bartolome 1981; Dyer et al. 1996).
Quantitative genetic variation in morphology and phenology exist among populations of N. pulchra, which evidently reflects adaptation to different environments (Huntsinger et al. 1996; Knapp & Rice 1998). In a common garden evaluation of 10 diverse natural populations of N. pulchra, quantitative genetic variation in plant height, culm length, per cent heading, leaf length and width, seeds/culm, seed weight and awn weight was found to be associated with climatic differences and geographical distance (Knapp & Rice 1998). Based on these results, it was suggested that use of nonlocal seed sources, translocated across different regions of California, could result in poorly adapted plantings that may ultimately fail (Knapp & Rice 1998). Moreover, patterns of isozyme variability among these 10 natural populations were correlated only with geographical distance and have limited usefulness in predicting the spatial scale over which N. pulchra seed should be translocated (Knapp & Rice 1998). Because measurements of quantitative trait variation are time- and labour-intensive, it was suggested that restorationists should use climatic information in making rapid decisions about appropriate spatial scales for translocating native grasses (Knapp & Rice 1998). The United States Department of Agriculture, Natural Resource Conservation Service, Plant Materials Center (PMC) at Lockeford, California has developed three natural germplasm sources of N. pulchra (e.g. LK115d, LK215e and LK315d). The identity of these natural N. pulchra germplasm sources reflect the Lockeford PMC as the releasing agency and the Major Land Resource Area (MLRA 15) for which these releases are best suited. Although MLRA boundaries were not intended to define seed transfer zones, these areas are characterized by particular patterns of soil type, climate, water resources and land use (USDA-SCS 1981). Approximately 50% of MLRA 15, Central California Coast Range, is native grass, shrub and brush rangeland. Another 35% is open woodland, also used for grazing. If plant cover is removed from the soils by fire, overgrazing, cultivation or logging, the hazard of erosion in this area is severe because of steep slopes and high-intensity rainfall (USDA-SCS 1981). Precipitation (typically 375–750 mm) in this area usually occurs during fall, winter or spring, but rarely in the summer. These natural germplasm sources (i.e. LK115d, LK215e and LK315d) were selected on the basis of flowering date, vigour, survival and growth from a replicated, common-garden evaluation of 32 accessions and released under Selected Class criteria of the Pre-Variety Germplasm Certification Standards (Association of Official Seed Certifying Agencies 1999; Young 1995). In order to protect the natural identity and diversity of these commercial germplasm sources, the foundation seed (used to establish commercial seed fields) was increased from seed collected directly from the original source populations. However, ecologists are also concerned that inadvertent directional selection may occur during cultivation (Knapp & Rice 1994). Moreover, determination of genetic identities for natural populations and commercial germplasm sources of native plant species is still a difficult problem of considerable importance to many public or private conservation groups and land managers.
The amplified fragment length polymorphism (AFLP) method (Vos et al. 1995) is a DNA fingerprinting technique that is highly informative in closely related germplasm (e.g. intraspecific) and can be used to estimate standard parameters of nucleotide sequence variation (π, DXY and DA) and differentiation among heterogeneous populations (GS). The extent of DNA polymorphism within a heterogeneous population is often measured by nucleotide diversity (π), which is defined as the average number of either nucleotide differences or substitutions per site for a group of DNA sequences (alleles) sampled (Nei & Li 1979; Nei & Miller 1990). Likewise, a standard measure of divergence between two heterogeneous populations is the net nucleotide divergence (DA), where the average π within populations has been subtracted from DXY, the total nucleotide divergence or average number of nucleotide differences per site between populations (Nei & Li 1979; Nei & Miller 1990). The apportionment of nucleotide sequence variation among (DA/DXY) populations is a standard measure of differentiation (GS). Nei & Li (1979) developed a method for estimating π and DXY using restriction endonucleases [restriction fragment length polymorphism (RFLP) method]. Variation within restriction/priming sites can also be used to estimate nucleotide sequence variation using the AFLP technique (Innan et al. 1999). However, a relatively small percentage (≈ 15–20%) of the AFLPs, analysed within genetic mapping populations of grasses, evidently arise from small insertion/deletions (indels) rather than nucleotide sequence variation at the restriction/priming sites (Becker et al. 1995; Bai et al. 1999; Larson et al. 2000b).
The primary objectives of this study were to determine mode of reproduction in N. pulchra and elucidate the genetic identities of LK115d, LK215e and LK315d relative to other natural germplasm sources collected within or near MLRA 15. Co-dominant AFLPs (i.e. indels) that vary within populations were identified and used to measure natural sefling rates (mode of reproduction). The remaining AFLP variation was used to estimate standard parameters of nucleotide sequence variation within and among populations. Differentiation among populations (GS) was measured by the partitioning of AFLP variation in terms of Shannon–Weaver phenotypic diversity values (within and among populations) and the proportion total nucleotide sequence variation (DXY) partitioned among populations (DA). The relationship between genetic distance and geographical distance was also investigated.
Materials and methods
Studies were initiated using foundation seed of LK115d, LK215e and LK315d and seed collected from each of 10 natural plants at 12 different sites (Table 1). Foundation seed of LK115d, LK215e and LK315d (lot numbered SFD-98-F2) was increased and certified under guidelines of the California Crop Improvement Association. Seed from the 12 natural populations was collected during April and May 1999. The spatial scale of these collection sites was approximately one ha or less. We attempted to germinate ≈ 150 seeds from each of the three foundation seed lots and 120 natural plants on moist blotter paper using a 2-week prechill at 5 °C, with the addition tetramethylthiuram disulfide as a fungicide. Seedlings were transplanted from germination boxes to individual cone soil containers maintained in an environmentally controlled greenhouse. Genomic DNA was extracted from ≈ 100 mg of young, green leaf tissue of each transplanted seedling, as described by Murray & Thompson (1980).
| County ID | (California) | Location | Latitude–Longitude |
|---|---|---|---|
| Tehama | Tehama | Red Bank Cr. | 40°03′54″–122°36′00″ |
| LK115d | Tehama | Hwy. 36 West of Bowman Rd. | 40°17′00″–122°29′00″ |
| Colusa | Colusa | Leesville | 39°09′51″–122°23′39″ |
| LK215e | Colusa | Walnut Valley Ranch, Lodoga | 39°20′00″–122°30′00″ |
| Calaveras | Calaveras | Hwy. 12 East of Evans Rd. | 38°11′04″–120°50′34″ |
| Contra Costa | Contra Costa | Mt. Diablo | 37°52′21″–121°51′22″ |
| Alameda | Alameda | Eden Canyon at I-580 | 37°42′00″–122°01′30″ |
| LK315d | Alameda | Rancho Los Mochos BSA Camp | 37°30′00″–121°32′00″ |
| Marin | Marin | Lagunitas Dam | 37°57′35″–122°36′36″ |
| SF Yerba Buena | San Francisco | Yerba Buena Island | 37°48′51″–122°21′41″ |
| SF Bermal | San Francisco | Bermal Hill | 37°44′36″–122°24′49″ |
| SF St. Mary | San Francisco | St. Mary’s Park | 37°43′45″–122°25′25″ |
| SF McLaren | San Francisco | John McLaren Park | 37°43′08″–122°24′59″ |
| SF Bayview | San Francisco | Bayview Hill | 37°42′57″–122°23′47″ |
| San Luis Obispo | San Luis Obispo | Waddell Ranch, Cayucos | 35°27′04″–120°57′04″ |
DNA fingerprinting was conducted using the AFLP technique according to the methods of Vos et al. (1995), except that EcoRI selective amplification primers were labelled with fluorescent 6-carboxy fluorescein (6-FAM) on the 5′ nucleotide. The AFLP adapter sequences, preamplification primer sequences and selective amplification primer sequences are described in Table 2. The amplified fragments were fractionated and detected with an ABI373XL instrument (PE Applied Biosystems, Foster City, CA, USA) using 34 cm well-to-read polyacrylamide gels formulated with 6.5% Long Ranger (FMC, Rockland, ME, USA), 7 m urea and 1× TBE running buffer. Each sample lane included the GS500-ROX (PE Applied Biosystems) internal lane size standards (labelled with fluorescent rhodamine X) ranging from 35 to 500 bp. Fluorescent signals of the 6-FAM-labelled AFLPs, between 35 and 500 bp, were identified using genescan 3.1 software (PE Applied Biosystems). The genescan sample (trace) files were subsequently analysed for the presence and absence of AFLP products, in ≈ 1 bp intervals, using Genographer (Benham et al. 1999). The degree of AFLP polymorphism and overall distribution of AFLP variation within and among populations was quantified using the Shannon–Weaver phenotypic diversity index as described for isozyme data (Chung et al. 1991; Dolan 1994; Knapp & Rice 1998) and AFLP data (Paul et al. 1997).
| Type | Name | Sequence |
|---|---|---|
| EcoRI adapter | 5′- CTCGTAGACTGCGTACC-3′ | |
| 3′- CATCTGACGCATGGTTAA-5′ | ||
| MseI adapter | 5′- GACGATGAGTCCTGAG-3′ | |
| 3′- TACTCAGGACTCAT-5′ | ||
| EcoRI + 1 primer | E01 | 5′- GACTGCGTACCAATTCA-3′ |
| EcoRI + 3 primers | E36 | 5′- GACTGCGTACCAATTCACC-3′ |
| E40 | 5′- GACTGCGTACCAATTCAGC-3′ | |
| E41 | 5′- GACTGCGTACCAATTCAGG-3′ | |
| MseI + 1 primer | M02 | 5′- GATGAGTCCTGAGTAAC-3′ |
| MseI + 3 primers | M47 | 5′- GATGAGTCCTGAGTAACAA-3′ |
| M48 | 5′- GATGAGTCCTGAGTAACAC-3′ | |
| M49 | 5′- GATGAGTCCTGAGTAACAG-3′ | |
| M50 | 5′- GATGAGTCCTGAGTAACAT-3′ | |
| M60 | 5′- GATGAGTCCTGAGTAACTC-3′ | |
| M62 | 5′- GATGAGTCCTGAGTAACTT-3′ |
To help identify allelic AFLP fragments (co-dominant markers), phi correlation coefficients (Φ) were calculated among all pair-wise comparisons of all fragments over all primer pairs, within primer pairs and among comparisons of fragments differing by 3 bp (within each primer pair) using SAS Version 8 (SAS Institute Inc., Cary, NC, USA). Allelic fragments that might arise from indels would be expected to show phi coefficients (Φ) equal to −1 among homozygous genotypes or ≈ −0.3 among genotypes in Hardy–Weinberg equilibrium (in a two allele system). The genotypic scores for all fragments differing by 3 bp (within each primer pair), where Φ < 0, were compared (Fig. 1) to identify co-dominant markers. These putative co-dominant markers were used to estimate selfing rates (S̄H), as described by Nordberg & Donnelly (1997) and Bergelson et al. (1998).

List of all AFLPs that display negative phi (Φ) coefficients (repulsion phase associations) in pairwise comparisons of fragments 3 bp different (within primer pairs). Fragments that appear to be allelic (i.e. putative co-dominant markers) are shown in bold text. Co-dominant markers that vary within these Nassella pulchra populations (highlighted text) were used to elucidate mode of reproduction. Populations ordered from left to right are as follows: (1) Tehama, (2) LK115d, (3) Colusa, (4) LK215e, (5) Calaveras, (6) Contra Costa, (7) Alameda, (8) LK315d, (9) Marin, (10) SF Yerba Buena, (11) SF St. Mary, (12) SF McLaren, (13) SF Bayview and (14) San Luis Obispo.
The computer program (Innan et al. 1999) used to estimate nucleotide variation based on F, the estimated proportion of shared AFLP fragments between haploid genomes (with allelic fragments treated as shared characters), was kindly provided by Hideki Innan (University of Tokyo, Japan). For inbred genotypes, these values of F are the same as the average genetic similarity coefficients between individual plants, compared within or among populations, as described by Nei & Li (1979). This method of estimating nucleotide variation is based on the fact that each AFLP product represents a 16-bp sequence assay when using the EcoRI and MseI restriction enzymes, which have 6 and 4 bp recognition sequences, respectively, and three selective nucleotides on each of the two AFLP selective amplification primers. Therefore, each shared AFLP product indicates zero nucleotide differences over 16 bp, whereas polymorphisms reflect at least one nucleotide difference over 16 bp. The actual number of differences that contribute to each polymorphism is a function of F, which can be used to determine the overall number of nucleotide substitutions per site. These methods for estimating the number of nucleotide differences based on the proportion of shared AFLP products are good when GC content is near 0.5 and the when the number of differences is not too large (Innan et al. 1999). This assumption is tentatively acceptable for Nassella pulchra because the GC contents of wheat and maize, two other divergent Poaceae species, are very similar (45.5 and 46.0%, respectively) and close to 50% (Salinas et al. 1988; Montero et al. 1992). Allele frequencies, similarity coefficients (Nei & Li 1979), and proportions of shared fragments were calculated with SAS.
A upgma cluster analysis and scaled dendogram were based on the proportion of shared bands (Nei & Li 1979) among individual genotypes (plants) developed using ntsys-pc, version 2.02 (Exeter Software, Setauket, NY, USA). Bootstrap confidence levels (BCL) were recovered from the 50% majority-rule consensus tree of 1000 upgma searches (replicates), using paup* Version 4.0b4a (Sinauer Associates, Inc., Sunderland, MA, USA). Matrix correspondence between genetic similarity (i.e. proportion of shared fragments) and geographical distance or nucleotide divergences (DXY or DA) and geographical distances among populations was also tested (Mantel 1967) using ntsys-pc.
Results
High germination rates and large vigorous seedlings were observed for the foundation seed lots of LK115d, LK215e and LK315d. However, some difficulties were encountered when germinating seed harvested from natural plants. The initial objective was to recover at least one seedling DNA sample from at least six natural plants at each of 12 collection sites (Table 1). However, only 86 of the 120 seed collections, harvested from 10 natural plants at each of 12 collection sites, produced at least one seedling. Only one viable seedling was recovered from the 10 seed collections harvested from the SF Bermal population (Table 1), therefore this population was omitted from the DNA analysis. Fewer than six of the natural plants sampled from the SF McLaren, SF St. Mary and Marin populations (Table 1) produced at least one viable seedling. Thus more than one seed from the same natural plant was used for the latter populations (Fig. 2). In general, poor caryopsis development and low seedling vigour were prevalent among seeds harvested from the natural plants. Although the LK115d, LK215e and LK315d natural germplasm sources were selected for vigour and other traits, some or most of the viability problems associated with seed collected from natural plants may be attributed to less favourable growing conditions. It is also likely that standard seed handling procedures would have removed poorly developed seeds from the foundation seed lots of LK115d, LK215e and LK315d.
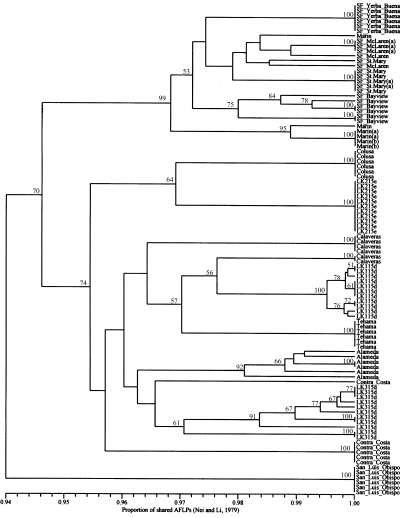
upgma phenogram of 98 Nassella pulchra genotypes from 11 natural populations and three Selected Class natural germplasm releases, based on 178 informative AFLPs. Seedling genotypes derived from the maternal plant(s) are annotated by identical letters in parentheses, i.e. SF_Mclaren(a), SF_St. Mary(a), Marin(a), and Marin(b); otherwise genotypes represent seedlings derived from different maternal plants. Bootstrap confidence levels are indicated for clades present in the 50% majority-rule consensus of 1000 upgma searches.
AFLP provided an efficient method of measuring nucleotide variation within and among the 14 Nassella pulchra populations. Eight AFLP primer pairs (E36M47, E36M48, E40M49, E40M50, E40M62, E41M47, E41M48, E41M60) amplified ≈ 407.6 fragments per plant (Table 3), or an average of ≈ 60 fragments per plant per primer pair. As expected, the number of AFLP fragments per plant was fairly consistent, ranging from 403 to 412 fragments per plant across the 98 plants examined (Table 3). The average size of these fragments was 240 nucleotides; with an average minimum size of 71 nucleotides and an average maximum size was 490 nucleotides. A total of 513 different fragments was resolved among these 98 DNA samples (Table 3), with an average of 64.1 fragments per primer pair. Nearly 36% of the fragments were polymorphic over all populations, whereas only 3.4% of the fragments were polymorphic within sample populations (Table 3). A total of 178 polymorphic fragments were detected including 125 fragments that varied within at least one population and 53 fragments that varied among, but not within, populations. The average Shannon–Weaver diversity (per AFLP primer pair) within populations (HS) was 0.60, whereas the total AFLP diversity (HT) was 5.77 (Table 3). Therefore, the proportion of total AFLP diversity among populations (GS) was 0.90, which is considerably greater than the partitioning of N. pulchra isozyme variation (GS = 0.57) using a similar Shannon–Weaver diversity index (Knapp & Rice 1998).
| Population | Number of plants | Average number of fragments per plant (range) | Number of different fragments | Polymorphic fragments (%) | Average Shannon-Weaver density (H) per AFLP primer pair |
|---|---|---|---|---|---|
| Tehama | 6 | 406 | 406 | 0 | 0.00 |
| (406) | (0.0) | ||||
| LK115d | 11 | 409.3 | 413 | 6 | 0.31 |
| (408–410) | (1.5) | ||||
| Calaveras | 6 | 407.3 | 419 | 23 | 1.33 |
| (407–408) | (5.5) | ||||
| Alameda | 6 | 408.5 | 420 | 23 | 1.17 |
| (407–410) | (5.5) | ||||
| LK315d | 11 | 407.6 | 428 | 38 | 1.63 |
| (406–410) | (8.9) | ||||
| Colusa | 6 | 404 | 404 | 0 | 0.00 |
| (404) | (0.0) | ||||
| LK215e | 11 | 411 | 411 | 0 | 0.00 |
| (411) | (0.0) | ||||
| Contra Costa | 6 | 407 | 424 | 32 | 0.40 |
| (407) | (7.8) | ||||
| SF St. Mary | 6 (5*) | 411 | 418 | 14 | 0.63 |
| (411) | (3.3) | ||||
| SF McLaren | 5 (3*) | 408 | 417 | 19 | 0.32 |
| (407–409) | (4.6) | ||||
| SF Bayview | 6 | 406.8 | 419 | 22 | 1.29 |
| (404–410) | (5.3) | ||||
| SF Yerba Buena | 6 | 412 | 412 | 0 | 0.00 |
| (412) | (0.0) | ||||
| Marin | 6 (4*) | 407.5 | 419 | 23 | 1.25 |
| (406–410) | (5.5) | ||||
| San Luis Obispo | 6 | 403 | 403 | 0 | 0.00 |
| (403) | (0.0) | ||||
| Overall | 98 | 407.6 | 513 | 178 | 5.77 |
| (403–412) | (35.7) |
- * Actual number of natural plants sampled without replication among (identical) progeny genotypes.
Small insertion/deletions (indels) were evident, particularly among comparisons of AFLPs 3 bp different (within primer pairs). The distributions of phi coefficients (Φ) among pair-wise comparisons of polymorphic AFLPs are almost identical when compared within and between primer pairs, except for the frequency of comparisons in which Φ = −1 (Table 4). The percentage of comparisons in which Φ = −1 is almost double within AFLP primer pairs compared with between AFLP primer pairs (Table 4). Nearly half (10) of these comparisons, in which Φ = −1, are among fragments 3 bp different (Table 4). Moreover, the percentage of comparisons in which Φ = −1 is almost 30 times greater among comparisons of fragments 3 bp different (within AFLP primer pairs), than the number of comparisons in which Φ = −1 between AFLP primer pairs (Table 4). Although 25% of the comparisons between AFLP primer pairs fall outside the 95% confidence interval (i.e. ≈ −0.20 to 0.20 with 98 plants) for zero association (Table 4), we do not recognize any molecular basis that would account for these correlations. We speculate that natural pedigree structure (relatedness within populations) effectively widens the 95% confidence interval for detecting significant associations between markers. In any case, we examined the possibility of indels among all pair-wise comparisons of fragments 3 bp different (within AFLP primer pairs) in which Φ < 0 (Fig. 1). Although larger indels may exist, small indels (i.e. 3 bp) evidently account for most of the unusual Φ coefficients observed within AFLP primer pairs (Table 4). In our interpretation of these data (Fig. 1), all indels show strong negative correlations (i.e. Φ = −1). Moreover, 12 of these 15 co-dominant markers (Fig. 1) show compelling evidence of high selfing rates (i.e. S̄H= 1) within populations, including Calaveras, Contra Costa, Alameda, LK315d, Marin, SF St. Mary, SF McLaren and SF Bayview. Little or no variation of any sort was detected within the other populations. Because, seedlings of different wild plants (or plants grown together in the foundation seed fields of LK315d) were analysed, we are reasonably certain that these results reflect genetic variation among true-breeding genotypes (selfing mode of reproduction) rather than variation among colonizing plants that originated from reproductively isolated populations. Homozygous genotypes within LK315d are particularly interesting considering that the parents of these seedlings were grown under conditions that should have been highly conducive to cross-fertilization.
| Phi (Φ) coefficients | Fragments between AFLP primer pairs (%) | Fragments within AFLP primer pairs (%) | Fragments ≤ 3 bp different within AFLP primer pairs (%) |
|---|---|---|---|
| = 1.00 | 85 (0.6) | 9 (0.5) | 0 (0.0) |
| 0.80–1.00 | 51 (0.4) | 10 (0.5) | 1 (1.8) |
| 0.60–0.80 | 141 (1.0) | 14 (0.7) | 0 (0.0) |
| 0.40–0.60 | 370 (2.7) | 45 (2.3) | 3 (5.4) |
| 0.20–0.40 | 1 170 (8.5) | 170 (8.7) | 6 (10.7) |
| 0.00–0.20 | 4 767 (34.5) | 676 (34.8) | 6 (10.7) |
| −0.20–0.00 | 5 395 (39.1) | 760 (39.1) | 17 (30.4) |
| −0.40–0.20 | 1 239 (9.0) | 165 (8.5) | 7 (12.5) |
| −0.60–0.40 | 327 (2.4) | 37 (1.9) | 2 (3.6) |
| −0.80–0.60 | 142 (1.0) | 20 (1.0) | 3 (5.4) |
| −1.00–0.80 | 41 (0.3) | 14 (0.7) | 1 (1.8) |
| = − 1.00 | 82 (0.6) | 23 (1.2) | 10 (17.9) |
| Number of comparisons | 13 810 | 1943 | 56 |
- AFLP, amplified fragment length polymorphism.
Although the size difference (3 bp) and frequency of putative allelic fragments (30/178) detected in this study are similar to that reported in AFLP studies of other grass (Becker et al. 1995; Bai et al. 1999; Larson et al. 2000b), it is possible that we failed to identify a substantial number of other indels. If we assume 30 fragments were allelic, then a total of 498 different, nonallelic sequences were analysed (i.e. 513 − 15 = 498). With an average size of 240 bp per fragment, this represents a sample of 119 520 bp. Virtually all of the indels within these sequences will produce polymorphic fragments. However, only 6.7% of the nucleotide variation within these sequences can be detected (i.e. 16 bp × 298 fragments = 7968 bp total). This inference follows from the fact that restriction/priming sites typically account for 16 bp per fragment, assuming one EcoRI site and one MseI site per fragment (Vos et al. 1995). Because five of these putative indels also showed evidence of nucleotide variation (Fig. 1), the actual number of polymorphisms detected was ≈ 173 (i.e. 178 polymorphic fragments −15 indels +5 compound polymorphisms) ignoring convergent mutations. Therefore, ≈ 8.7% of the polymorphisms were indels and the remaining 91.3% were nucleotide substitutions at the restriction/priming sites. Because only 6.7% of the nucleotide sequence variation can be detected using the AFLP method, described by Vos et al. (1995), we deduce that nucleotide substitutions were ≈ 157-fold more common than indels. Bergelson et al. (1998) demonstrate that the frequency of nucleotide substitutions is sevenfold more common than indels in a survey of 5346 bp divided among coding and noncoding sequences of the Adh, Dhs1 and Gpa1 loci in Arabidopis thaliana. Although it is possible that we failed to identify a substantial number of indels, our estimate of selfing rate (i.e. S̄H= 1) in N. pulchra is not likely to change in any case.
The average proportion of shared AFLP fragments (F) within and among populations was 0.995 and 0.953, respectively (Table 5). However, to estimate nucleotide sequence variation within and among populations, corrected values for the average proportion of shared AFLP fragments (F) were also computed by treating allelic fragments as shared characters (Table 5). The average proportion of shared AFLP fragments (F), corrected for indels, within and among populations was 0.996 and 0.962, respectively. As expected, values of F are higher when corrected for indels. The corresponding estimates of nucleotide variation within (π) and among populations (DXY) were 0.00035 and 0.00247, respectively. Accordingly, the net nucleotide divergence among populations (DA) is 0.00212 and the proportion of total nucleotide variation among populations (GS) is 0.86. Hence, the partitioning of nucleotide variation (GS = 0.86) among these sample populations of N. pulchra is somewhat less than differentiation of among the Shannon–Weaver diversity values for AFLP variation (GS = 0.90), but still much greater than the partitioning of Shannon–Weaver values (GS = 0.57) for isozyme variation (Knapp & Rice 1998). Interestingly, estimates of nucleotide diversity (π = 0.0004), total nucleotide divergence (DXY = 0.0014) and differentiation (GS = 0.64) among populations of self-fertilizing A. thaliana (Bergelson et al. 1998) are very similar to our estimates for N. pulchra. Although population differentiation is certainly high in N. pulchra, estimates of nucleotide variation may still be substantially inflated by undetected indels.
| Teham. (1) | LK115d (2) | Colusa (3) | LK215e (4) | Calav. (5) | Contra Costa (6) | Alam. (7) | LK315d (8) | Marin (9) | SF Yerba Buena (10) | SF St. Mary (11) | SF McLar (12) | SF Bay (13) | San Luis Obispo (14) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (1) | 1.000 | |||||||||||||
| (1.000) | ||||||||||||||
| (2) | 0.970 | 0.998 | ||||||||||||
| (0.977) | (0.998) | |||||||||||||
| (3) | 0.956 | 0.963 | 1.000 | |||||||||||
| (0.965) | (0.971) | (1.000) | ||||||||||||
| (4) | 0.950 | 0.953 | 0.969 | 1.000 | ||||||||||
| (0.962) | (.963) | (0.977) | (1.000) | |||||||||||
| (5) | 0.966 | 0.968 | 0.958 | 0.950 | 0.989 | |||||||||
| (0.970) | (.979) | (0.972) | (0.965) | (0.992) | ||||||||||
| (6) | 0.957 | 0.957 | 0.958 | 0.943 | 0.962 | 0.991 | ||||||||
| (0.967) | (0.971) | (0.972) | (0.959) | (0.973) | (0.995) | |||||||||
| (7) | 0.958 | 0.963 | 0.965 | 0.951 | 0.960 | 0.954 | 0.991 | |||||||
| (0.965) | (0.968) | (0.971) | (0.960) | (0.971) | (0.970) | (0.992) | ||||||||
| (8) | 0.957 | 0.964 | 0.962 | 0.955 | 0.958 | 0.958 | 0.963 | 0.987 | ||||||
| (0.966) | (0.970) | (0.972) | (0.966) | (0.971) | (0.972 | (0.969) | (0.989) | |||||||
| (9) | 0.948 | 0.944 | 0.949 | 0.944 | 0.948 | 0.949 | 0.949 | 0.943 | 0.991 | |||||
| (0.955) | (0.953) | (0.956) | (0.949) | (0.958) | (0.961) | (0.957) | (0.955) | (0.992) | ||||||
| (10) | 0.941 | 0.939 | 0.946 | 0.950 | 0.958 | 0.946 | 0.947 | 0.943 | 0.967 | 1.000 | ||||
| (0.954) | (0.954) | (0.958) | (0.955) | (0.959) | (0.962) | (0.960) | (0.959) | (0.974) | (1.000) | |||||
| (11) | 0.945 | 0.946 | 0.950 | 0.945 | 0.950 | 0.947 | 0.945 | 0.943 | 0.969 | 0.975 | 0.995 | |||
| (0.951) | (0.956) | (0.958) | (0.955) | (0.959) | (0.958) | (0.955) | (0.955) | (0.975) | (0.980) | (0.996) | ||||
| (12) | 0.948 | 0.944 | 0.950 | 0.949 | 0.950 | 0.948 | 0.947 | 0.942 | 0.973 | 0.973 | 0.980 | 0.992 | ||
| (0.957) | (0.958) | (0.961) | (0.958) | (0.962) | (0.961) | (0.960) | (0.958) | (0.977) | (0.979) | (0.983) | (0.993) | |||
| (13) | 0.945 | 0.947 | 0.952 | 0.950 | 0.950 | 0.953 | 0.949 | 0.947 | 0.971 | 0.971 | 0.973 | 0.973 | 0.990 | |
| (0.952) | (0.955) | (0.960) | (0.956) | (0.960) | (0.964) | (0.959) | (0.958) | (0.973) | (0.977) | (0.978) | (0.973) | (0.991) | ||
| (14) | 0.937 | 0.945 | 0.947 | 0.936 | 0.938 | 0.938 | 0.944 | 0.936 | 0.941 | 0.935 | 0.944 | 0.939 | 0.946 | 1.000 |
| (0.944) | (0.947) | (0.949) | (0.941) | (0.949) | (0.947) | (0.948) | (0.943) | (0.945) | (0.945) | (0.949) | (0.948) | (0.951) | (1.000) |
In most cases, cluster analysis, based on the proportion of shared AFLP fragments between individual seedlings (Table 5), grouped members of each population together (Fig. 2). Moreover, bootstrap confidence levels indicate that most natural populations (e.g. SF_Yerba Buena, SF Bayview, Colusa, LK215e, LK115d, Tehama, Alameda, LK315d and San Luis Obispo) can be distinguished unambiguously from each other using the AFLP technique (Fig. 2). Geographical distance among original collection sites (Table 1) was highly correlated (P ≤ 0.01) with genetic variation among these sample populations. Geographical distance was significantly (P = 0.0020) correlated (r = 0.60) with DXY. Likewise, geographical distance was significantly (P = 0.0030) correlated (r = 0.55) with DA, albeit less than DXY. As would be expected LK115d, LK215e and LK315d were generally most similar to natural populations (i.e. Tehama, Colusa, Alameda) collected from sites near the respective source populations of these Selected Class natural germplasm accessions (Fig. 2). Interestingly, the LK315d population, collected from Alameda Co., displayed considerably more AFLP diversity than LK115d or LK215e (Table 3, Fig. 2). Like LK315d, the natural population collected from Alameda Co. was also more diverse than most of the other populations.
Although geographical distance and genetic variation were significantly correlated, cluster analysis of the Calaveras, Marin and SF McLaren seedlings (Fig. 2) reveals mixtures of true-breeding genotypes that transcend and belie geographical origin. These observations may reflect seed dispersal from genetically different populations. Interestingly, the Contra Costa, Alameda and LK315 populations, from the inland side of San Francisco Bay, show a stronger affinity to other inland populations (i.e. Colusa, LK215e, Calaveras, LK115d and Tehama) that originate from collection sites much further away compared with populations collected from the nearby coastal side of San Francisco Bay (i.e. SF Yerba Buena, SF McLaren, SF St. Mary, SF Bayview and Marin). This putative division of coastal and inland populations, across San Francisco Bay was supported by the bootstrap confidence intervals (Fig. 2). Hierarchical analysis of DXY values (Table 6) also indicates substantial differentiation (GS = 0.30) between these two groups. Knapp & Rice (1998) assert that quantitative morphological variation among N. pulchra populations is primarily determined by climatic differences between the interior and coastal or maritime regions.
| Teham. (1) | LK115d (2) | Colusa (3) | LK215e (4) | Calav. (5) | Contra Costa (6) | Alam. (7) | LK315d (8) | Marin (9) | SF Yerba Buena (10) | SF St. Mary (11) | SF McLar (12) | SF Bay (13) | San Luis Obispo (14) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (1) | 0.00 | 0.85 | 2.23 | 2.43 | 1.66 | 1.95 | 1.98 | 1.83 | 2.64 | 2.95 | 3.03 | 2.54 | 2.81 | 3.62 |
| (2) | 1.45 | 1.20 | 1.31 | 1.76 | 0.48 | 1.09 | 1.19 | 0.97 | 2.17 | 2.35 | 2.10 | 2.00 | 2.01 | 2.82 |
| (3) | 2.23 | 1.91 | 0.00 | 1.45 | 1.53 | 1.63 | 1.59 | 1.44 | 2.57 | 2.82 | 2.70 | 2.27 | 2.28 | 3.29 |
| (4) | 2.43 | 2.36 | 1.45 | 0.00 | 1.98 | 2.47 | 2.31 | 1.83 | 3.04 | 2.89 | 2.77 | 2.60 | 2.54 | 3.82 |
| (5) | 1.91 | 1.33 | 1.78 | 2.23 | 0.50 | 1.31 | 1.34 | 1.25 | 2.32 | 2.37 | 2.25 | 1.96 | 2.03 | 3.04 |
| (6) | 2.10 | 1.84 | 1.78 | 2.62 | 1.71 | 0.31 | 1.51 | 1.28 | 2.09 | 2.28 | 2.41 | 2.12 | 1.87 | 3.27 |
| (7) | 2.23 | 2.04 | 1.84 | 2.56 | 1.84 | 1.91 | 0.50 | 1.38 | 2.26 | 2.31 | 2.52 | 2.09 | 2.09 | 3.10 |
| (8) | 2.17 | 1.91 | 1.78 | 2.17 | 1.84 | 1.78 | 1.97 | 0.69 | 2.30 | 2.28 | 2.42 | 2.26 | 2.20 | 3.35 |
| (9) | 2.89 | 3.02 | 2.82 | 3.29 | 2.82 | 2.49 | 2.76 | 2.89 | 0.50 | 1.40 | 1.21 | 0.98 | 1.18 | 3.30 |
| (10) | 2.95 | 2.95 | 2.82 | 2.89 | 2.62 | 2.43 | 2.56 | 2.62 | 1.65 | 0.00 | 1.14 | 1.11 | 1.17 | 3.55 |
| (11) | 3.15 | 2.82 | 2.82 | 2.89 | 2.62 | 2.69 | 2.89 | 2.89 | 1.58 | 1.26 | 0.25 | 0.73 | 0.99 | 3.17 |
| (12) | 2.76 | 2.82 | 2.49 | 2.82 | 2.43 | 2.49 | 2.56 | 2.82 | 1.45 | 1.33 | 1.07 | 0.44 | 0.89 | 3.13 |
| (13) | 3.09 | 2.89 | 2.56 | 2.82 | 2.56 | 2.30 | 2.62 | 2.82 | 1.71 | 1.45 | 1.39 | 1.39 | 0.56 | 2.87 |
| (14) | 3.62 | 3.42 | 3.29 | 3.82 | 3.29 | 3.42 | 3.35 | 3.69 | 3.55 | 3.55 | 3.29 | 3.35 | 3.15 | 0.00 |
Discussion
Stebbins (1957) suggests that self-fertilization, in grasses and other plants, may be advantageous: (i) when conditions are unfavourable for crossing (such as periodic drought on the annual grasslands of California); (ii) for reproduction of one or two colonists following long-distance dispersal; and (iii) for reproduction of ‘weedy’ genotypes that are well-adapted to the disturbed niche, in which populations are constantly destroyed and quickly built up again. Indeed, evidence suggests (Bartolome 1981; Bartolome & Gemmill 1981) that Nassella pulchra is an ephemeral species that has evolved to rapidly colonize or increase abundance on many disturbed sites. Results of this study indicate that self-fertilization in N. pulchra is another important adaptation that may be related to dispersal and colonization by this species.
Self-fertilization in N. pulchra has reduced variation within populations and increased apparent differences among populations. Estimates of nucleotide diversity within self-fertilizing populations of N. pulchra (0.00035) are almost two orders of magnitude less than estimates of nucleotide diversity within cross-fertilizing cultivars of Psuedoroegneria spicata (0.03400), also determined using the AFLP method (Larson et al. 2000a). Moreover, estimates of total nucleotide diversity within N. pulchra (0.00247) are also considerably less the estimates of nucleotide diversity in a multiple-origin polycross of 25 P. spicata populations (0.0366). Although no attempt was made to distinguish indels from nucleotide substitutions in P. spicata (Larson et al. 2000a), substantially more AFLP variation was observed in P. spicata than N. pulchra. Conversely, estimates of nucleotide differentiation within N. pulchra (GS = 0.86) are more than one order of magnitude larger than differentiation between cultivars of P. spicata (GS = 0.07). Large reductions in genetic variability within populations and increased differentiation among populations have also been observed in other self-fertilizing grasses (Price et al. 1984). Large values of GS generally indicate restricted gene flow between populations (Bergelson et al. 1998), which in most plants occurs by pollen flow and/or seed dispersal. Although pollen flow is obviously reduced or eliminated in self-fertilizing species, seed dispersal of N. pulchra is probably equal to or greater than seed dispersal of P. spicata. Selection and clonal expansion may also influence GS by reducing variability within self-fertilizing populations (Bergelson et al. 1998). Clonal expansion of inbred founders may be expected to produce patches of homogeneous populations for weedy ephemeral species such as Arabidopsis thaliana (Bergelson et al. 1998). Relative to a cross-fertilizing population, selfing per se is only expected to reduce nucleotide variation by a factor of two when the selfing rate (S̄H) approaches one (Pollack 1987). Because recombination is reduced by self-fertilization, selection can also reduce the neutral genetic variability across large linkage blocks of DNA by way of hitchhiking effects or selective sweeps associated with favourable genes (Maynard Smith & Haigh 1974) or background selection against deleterious mutations (Charlesworth et al. 1993). Assuming normal rates of deleterious mutations, background selection may be expected to reduce the accumulation of neutral genetic variability within populations by another factor of 10 in a self-fertilizing population (Charlesworth et al. 1993). Therefore, a number of factors including reduced gene flow, selection and clonal expansion of inbred founders, may have reduced variation within self-fertilizing populations of N. pulchra and increased differentiation among these demes.
AFLP variation may reflect neutral genetic differences among populations of N. pulchra, not evident by isozyme analysis. In common garden studies, Knapp & Rice (1998) demonstrate that quantitative morphological variation among N. pulchra populations is correlated (P = 0.0142) with climatic differences across a relatively narrow range of longitudes, whereas isozyme variation is correlated (P = 0.0036) with geographical distance across a relatively wide range of latitudes. Based on these results, Knapp & Rice (1998) assert that poorly adapted plantings may result when using seed transferred between coastal and inland sites. Moreover, Knapp & Rice (1998) allege that isozymes (and perhaps molecular markers in general) have limited usefulness in predicting adaptive differences. However, the lack of congruence between morphological variation and isozyme variation (Knapp & Rice 1998) may reflect special limitations of the isozyme surveys that do not apply to methods of DNA fingerprinting that are potentially more informative. Isozymes are suitable for screening a relatively small number of neutral characters in a large number of plants (per population). However, estimates of genetic diversity and distance are affected far more severely by the number of loci sampled than by the number of individuals sampled (Gorman & Renzi 1979; Nei 1987). Even a few representative individuals may be sufficient to measure DNA variation if the number of loci examined is large. Indeed, the partitioning of Shannon–Weaver diversity values for AFLPs (GS = 0.90), based on a relatively large number of loci in fewer individuals, is considerably different from the partitioning of isozyme banding patters (GS = 0.57), even though the populations surveyed for isozyme variation originate from much wider range of latitudes. Although AFLP variation is also correlated with geographical distance, we observed a discontinuity of AFLP variation across a narrow region (San Francisco Bay and adjoining coastal mountains) separating coastal and inland areas that may be related to putative differences between the inland and coastal regions (Knapp & Rice 1998). Considering the self-fertilizing mode of reproduction and almost complete partitioning of all genetic variation among these populations of N. pulchra, it seems very likely that selection has somehow shaped AFLP variation in this species. Conversely, if genetic drift accounts entirely for the partitioning of AFLP variation among populations of N. pulchra then it is also likely that genetic drift had profound effects on the partitioning of quantitative morphological variation. We speculate that both selection and genetic drift influenced the partitioning of AFLP variation and quantitative morphological differences among natural populations of N. pulchra. Although adaptive variation within and among populations of N. pulchra may be important (Knapp & Rice 1998), there is not yet enough evidence to support the clear distinction or identification of two (i.e. inland and coastal) or more regionally distinct forms or ecotypes of N. pulchra. More extensive sampling of different sites is needed to detect genetic divergence among regionally distinct or ecologically distinct forms of N. pulchra. Hierarchical geographical analysis of genetic differences, within self-fertilizing species such as N. pulchra, might be efficiently performed by DNA fingerprinting a small number of plants from each of many different sites, perhaps even just one plant per site.
As expected, foundation seed lots of LK115d, LK215e and LK315d were genetically well defined, naturally diverse and most related to natural populations collected from nearby sites. Therefore, cultivated seed increases of LK115d, LK215e and LK315d probably did not have any inadvertent consequences on the genetic integrity of these commercial germplasm sources. Based on results of this study, we do not think that these plant materials are likely to cross with natural populations. However, the spatial scale of genetic identity among these Selected Class natural germplasm accessions and other natural populations is evidently very small. Moreover, the genetic identity of LK115d, LK215e and LK315d did not completely match that of natural populations collected from sites near the original source of these Selected Class germplasm releases. In fact, the AFLP method exposed genetic differences among virtually all of the natural populations, even within the city of San Francisco. Yet, one of the most ecologically important attributes of N. pulchra may be its ability to colonize disturbed sites (Bartolome 1981; Bartolome & Gemmill 1981). Moreover, much of the obvious variation among populations of N. pulchra may be the result of inbreeding and genetic drift associated with clonal expansion of inbred founders. Of course, the value of genetic diversity and importance of germplasm conservation are widely recognized and greatly appreciated. To ensure planting success and adaptation, more than one commercial germplasm source (e.g. LK115d, LK215e and LK315d) could be used in the same planting. Ideally, native plant workers could develop and use genetically diverse synthetics (cross-fertilizing species) or composites (self-fertilizing species) for each region or ecotype, where in fact these intraspecific differences among regions or ecotypes may be significant. However, morphological variation would also be problematic for seed production, particularly for composites of true-breeding genotypes. In any case, the motive to use native species in large-scale revegetation efforts needs to be complemented by effective germplasm utilization, innovative agronomic techniques, and a viable industry that can efficiently produce a readily available source of high-quality seed for these species. Thus, commercial germplasm of native grass species can be made practically available and useful in many conservation plantings.
Acknowledgements
This work was supported by joint contributions of the USDA, the Department of Defense Strategic Environmental Research and Development Program CS1103 project, and the US Army BT25-EC-B09 project. We also acknowledge the assistance of Dr Eric Knapp, Dr Jeffery Steiner, and journal referees in providing helpful reviews of this manuscript.
References
The USDA-NRCS Plant Materials Program aims to identify and develop a wide diversity of plant materials to solve environmental and conservation problems across all regions of the United States. The USDA-ARS Forage and Range Research Laboratory performs applied research related to many pasture and rangeland plants important to the western United States.




