Population structure in two sympatric species of the Lake Tanganyika cichlid tribe Eretmodini: evidence for introgression
Abstract
Patterns of genetic differentiation were analysed and compared in two sympatric species of the endemic Lake Tanganyika cichlid tribe Eretmodini by means of mitochondrial DNA (mtDNA) sequences of the control region and six microsatellite DNA loci. The sample area covers a total of 138 km of mostly uninterrupted rocky shoreline in the Democratic Republic of Congo and includes the entire distribution range of Tanganicodus cf. irsacae that stretches over a distance of 35 km. Both markers detected significant genetic differentiation within and between the two species. T. cf. irsacae contained lower overall genetic variation than Eretmoduscyanostictus, possibly due to its more restricted range of distribution and its smaller effective population sizes. Complete fixation of Tanganicodus mtDNA haplotypes was observed in Eretmodus at two localities, while at two other localities some Tanganicodus individuals possessed Eretmodus mtDNA haplotypes. Taking into account the relatively large average sequence divergence of 6.2% between the two species, as well as the geographical distribution of mtDNA haplotypes in the lake, the observed pattern is more likely to be a consequence of asymmetric introgression than of shared ancestral polymorphism. As there is significant population differentiation between sympatric Tanganicodus and Eretmodus populations, the events of introgressions may have happened after secondary contact, but our data provide no evidence for ongoing gene flow and suggest that both species are reproductively isolated at present time.
Introduction
The cichlid species flocks of the East African Great Lakes provide excellent opportunities to study speciation and adaptive radiation (Fryer & Illes 1972; Meyer 1993). Several cichlid lineages show unique levels of ecological diversification and species packing, and exceptionally fast speciation rates have been reported for the haplochromine lineages in lakes Malawi and Victoria (Seehausen et al. 1999). Each species flock is believed to have evolved independently from different ancestral riverine stocks via intralacustrine speciation (Meyer 1993). As lakes Tanganyika, Malawi, and Victoria differ widely in their relative ages, they offer a unique opportunity to study different stages of adaptive radiations in cichlids and to detect general and unique characteristics of their species flocks (Sturmbauer 1998).
Among several potential mechanisms promoting speciation in lacustrine cichlids, disruptive sexual selection on polymorphic male colouration has been suggested to play an important role during sympatric speciation in polygynous, maternal mouthbrooders, such as the haplochromines from lakes Malawi and Victoria (Seehausen et al. 1999). Other studies indicate that selection for ecological divergence may also be a major driving force of cichlid speciation (e.g. Schliewen et al. 1994). The evolution of divergent phenotypes and resource use may precede or initiate the speciation process, or it may evolve after reproductive isolation via disruptive sexual selection has occurred, and thus would facilitate the coexistence of species (Galis & Metz 1998). Other hypotheses for speciation in rock-dwelling cichlids emphasize the importance of extrinsic factors for speciation, such as lake level fluctuations promoting geographical isolation (Fryer & Illes 1972; Ribbink et al. 1983).
With an estimated age of 9–12 Myr Lake Tanganyika is the oldest of the East African Rift Lakes (Cohen et al. 1993) harbouring approximately 200 endemic cichlid species. In sharp contrast to the species flocks of haplochromine cichlids of lakes Malawi and Victoria, Lake Tanganyika contains several evolutionary lineages that probably predate the lake’s ancient history (Nishida 1991; Sturmbauer & Meyer 1993). Past lake level drops, some of which have split the lake up into isolated subbasins, have strongly affected rocky habitat patches in the littoral of Lake Tanganyika (Cohen et al. 1997). The effects of such changes in water level, at different temporal and spatial scales, were early hypothesized by Brooks (1950) and Poll (1952) to be one of the underlying causes of the faunal diversity found in Lake Tanganyika (but see Snoeks et al. 1994). Recently, molecular studies confirmed that these major vicariant events strongly influenced the phylogeographic patterns in some rock-dwelling cichlids (Sturmbauer & Meyer 1992; Sturmbauer et al. 1997, 2001; Rüber et al. 1999; but see Meyer et al. 1996). In Lake Malawi, which has persisted as a single basin throughout its history, several haplochromine species exhibit high levels of population subdivision, which points to the importance of allopatric divergence among habitat patches for speciation of the stenotopic mbuna cichlids that do not disperse over areas of deep water and/or long stretches of sandy shoreline (van Oppen et al. 1997a; Arnegard et al. 1999; Markert et al. 1999). The existence of concordant patterns of population structure across sympatric taxa would be a strong indication of the importance of shared historical factors for shaping population demography. On the other hand, noncongruent patterns may help to identify important differences among taxa concerning their dispersal abilities and habitat requirements that are relevant for past demographic events and evolutionary processes.
Cichlids from the tribe Eretmodini are endemic to Lake Tanganyika and occur along shallow rocky and pebble shorelines, mostly at depths lower than 5 m. A recent phylogenetic analysis based on mitochondrial DNA (mtDNA) sequences identified six distinct eretmodine lineages which are not concordant with the current taxonomy and suggest the ‘existence’ of cryptic species (Rüber et al. 1999). Based on the mtDNA phylogeny, the genera Eretmodus, Spathodus, and Tanganicodus are considered nonmonophyletic and are thus in need of a taxonomic reassessment. A lake-wide phylogeographic study demonstrated a high degree of intralacustrine endemism of the eretmodine lineages (Rüber et al. 1999). It was suggested that both the patchy distribution of rocky shorelines and Pleistocene lake level fluctuations have significantly influenced distribution patterns in these cichlids. Interestingly, similar trophic phenotypes assigned to different genetic lineages generally show complementary distributions. This observation was taken as evidence that species interactions might have also played a determinant role in shaping their present distribution (Rüber et al. 1999).
The two nominal species Eretmodus cyanostictus and Tanganicodus cf. irsacae (defined as mtDNA lineage C and D in Rüber et al. 1999) are easily distinguishable by differences in their body shape, dentition and colouration (Huysseune et al. 1999; Rüber et al. 1999; Rüber & Adams 2001). Eretmodus is an algae scraper, whereas Tanganicodus is a specialized invertebrate picker (Yamaoka et al. 1986). The two species are monogamous, sexually monochromatic, biparental mouthbrooders that perform a female-to-male shift of the hatchlings during mouthbrooding (Kuwamura et al. 1989). T. cf. irsacae has the most narrow distribution of all eretmodine cichlids and is only known from four localities over a distance of about 35 km in the south-western part of Lake Tanganyika in the Democratic Republic of Congo (Fig. 1). E. cyanostictus, on the other hand, has a much wider distribution range that covers about 600 km of coastline in the southern part of the lake.
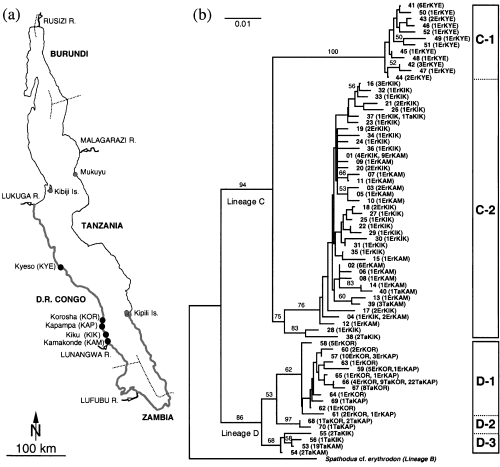
(a) Map of Lake Tanganyika with localities from where fish were collected for this study. Locality abbreviation: Kyeso (KYE), Kiku (KIK), Kapampa (KAP), Korosha (KOR), and Kamakonde (KAM). Known distribution of Eretmodus cyanostictus is indicated in grey. The distribution of Tanganicodus cf. irsacae is limited to the coastline between KIK and KAM. (b) Neighbour-joining tree based on partial control region sequences of the 70 haplotypes found among 198 sequenced specimens. Haplotype numbers (see Appendix II) are given next to each branch; in brackets, the number of individuals, species assignment (Er =Eretmodus cyanostictus; Ta =Tanganicodus cf. irsacae), and localities are given. The tree is rooted with Spathodus cf. erythrodon. Bootstrap percentage values based on 1000 replications are shown above branches. The assignments to the major genetic lineages (C and D) and clades are given in boxes.
In this paper we characterize the patterns of genetic variation in these two cichlid species, at localities where they are sympatric, using microsatellite markers and sequences of the mtDNA control region. The objectives of this study are: (i) to examine differences in genetic diversity and population structure that may be related to differences in distribution ranges; and (ii) to investigate the possibility of introgression between these closely related species under sympatric conditions.
Material and methods
Sample collection
A total of 123 specimens of Eretmoduscyanostictus were collected from five localities covering 138 km of mostly uninterrupted rocky shoreline, and 75 specimens from Tanganicodus cf. irsacae were collected from four localities in the Democratic Republic of Congo (Fig. 1), covering its entire known distribution range of 35 km. This study includes two populations of Eretmodus (KOR and KAP; for locality abbreviations see Fig. 1) that were not studied in a previous phylogenetic analysis (Rüber et al. 1999). For the microsatellite analyses two populations (Eretmodus KAP and Tanganicodus KIK) were omitted due to small sample sizes, therefore, a total of 117 specimens of E.cyanostictus and 69 specimens of T. cf. irsacae were used. Samples were preserved in 70% ethanol and voucher specimens have been deposited in the Africa Museum in Tervuren, Belgium.
Characterization of microsatellite DNA
DNA was extracted using a proteinase K, phenol–chloroform protocol (Kocher et al. 1989). A total of 10 microsatellite loci, which were developed for other East African cichlid species, were initially tested by sequencing to assess their utility for population studies in eretmodine cichlids. For sequencing, polymerase chain reactions (PCR) were carried out in 25 µL volumes [Tris 10 mm, pH 9.0, 1.5 mm MgCl2, 0.2 mm of each dNTP, 0.2 µm of each primer, and 0.65 units of Taq DNA polymerase (Pharmacia)] using the PCR conditions given in Kellog et al. (1995) for UNH001 and UNH002, Zardoya et al. (1996) for TmoM5, TmoM7, TmoM11, TmoM13, TmoM25, and TmoM27, and van Oppen et al. (1997b) for Pzeb1 and Pzeb3, with adjustments of annealing temperatures (Table 1). PCR fragments were either ligated into pGEM-T plasmid vectors (Promega) or using the TopoTA cloning kit (Invitrogene). Clones were sequenced with M13 universal (−40) and reverse primers with an Applied Biosystem 373 A DNA sequencer using the Taq Dye Deoxy Terminator Cycle Sequencing Kit FS (PE Biosystems).
| Locus | Specimen | Core sequence | Length | T a (°C) | Reference |
|---|---|---|---|---|---|
| TmoM5 * | Tropheus moorii | ( GC)6( AC)14 | 55 | Zardoya et al. (1996) | |
| Spathodus cf. erythrodon (B) | ( AC)33 | 314 | (AJ407057) | ||
| Eretmodus cyanostictus (C) | ( GC) 4( AC)13 | 282 | (AJ407058) | ||
| TmoM11 | Tropheus moorii | ( AC)19 | 50 | Zardoya et al. (1996) | |
| Eretmodus cf. cyanostictus (A) | ( AC)16 | 169 | (AJ407063) | ||
| Eretmodus cyanostictus (C) | ( AC)10 | 157 | (AJ407062) | ||
| Tanganicodus cf. irsacae (E) | ( AC)20 | 177 | (AJ407064) | ||
| TmoM25 | Tropheus moorii | ( CA)23 | 50 | Zardoya et al. (1996) | |
| Tanganicodus irsacae (A) | ( CA)5CGT( CA)28 | 374 | (AJ407069) | ||
| Eretmodus cf. cyanostictus (A) | ( CA)6CGT( CA)14 | 348 | (AJ407068) | ||
| Eretmodus cyanostictus (C) | ( CA)5CGT( CA)12 | 342 | (AJ407067) | ||
| Tanganicodus cf. irsacae (E) | ( CA)5CGT( CA)22 | 362 | (AJ407070) | ||
| Pzeb1 | Pseudotropheus zebra | ( GT)59CC( T)16 | 55 | van Oppen et al. (1997b) | |
| Eretmodus cyanostictus (C) | ( GT)17GA( T)13 | 140 | (AJ407052) | ||
| Tanganicodus cf. irsacae (D) | ( GT)7TTGC( T)15 | 124 | (AJ407053) | ||
| Pzeb3 | Pseudotropheus zebra | ( TG)12 | 55 | van Oppen et al. (1997b) | |
| Eretmodus cyanostictus (C) | ( TG)10 | 315 | (AJ407056) | ||
| Eretmodus cyanostictus (C) | ( TG)10 | 315 | (AJ407055) | ||
| Tanganicodus cf. irsacae (D) | ( TG)9 | 313 | (AJ407054) | ||
| UNH002 | Melanochromis‘chipokae’ | ( CA)23 | 52 | Kellog et al. (1995) | |
| Tanganicodus irsacae (A) | ( CA)13 | 162 | (AJ407076) | ||
| Spathodus cf. erythrodon (B) | ( CA)14 | 164 | (AJ407075) | ||
| Eretmodus cyanostictus (C) | ( CA)15 | 166 | (AJ407077) | ||
| TmoM7 | Tropheus moorii | ( CA) AA( CA)10 | 48 | Zardoya et al. (1996) | |
| Eretmodus cyanostictus (C) | ( CA) AA( CA)5 | 310 | (AJ407059) | ||
| Tanganicodus cf. irsacae (E) | ( CA) AA( CA)6 | 312 | (AJ407060) | ||
| Tanganicodus cf. irsacae (E) | ( CA) AA( CA)6 | 312 | (AJ407061) | ||
| TmoM13 | Tropheus moorii | ( CA)4CG( CG)24 | 50 | Zardoya et al. (1996) | |
| Spathodus cf. erythrodon (B) | ( CA)8 | 201 | (AJ407066) | ||
| Eretmodus cyanostictus (C) | ( CA)7 | 199 | (AJ407065) | ||
| TmoM27† | Tropheus moorii | ( CA)13CC( CA)5 | 55 | Zardoya et al. (1996) | |
| Eretmodus cyanostictus (C) | ( CA)5CC( CA)9 | 180 | (AJ407050) | ||
| Tanganicodus cf. irsacae (D) | ( CA)5CC( CA)7 | 176 | (AJ407051) | ||
| UNH001 | Melanochromis‘chipokae’ | ( AG)5–19 bp-( AC)35 | 52 | Kellog et al. (1995) | |
| Tanganicodus irsacae (A) | ( AG)5–19 bp-( AC)8 | 144 | (AJ407074) | ||
| Eretmodus cyanostictus (C) | ( AG)5–19 bp-( AC)7 | 142 | (AJ407072) | ||
| Eretmodus cyanostictus (C) | ( AG)5–19 bp-( AC)7 | 142 | (AJ407073) | ||
| Tanganicodus cf. irsacae (E) | ( AG)5–19 bp-( AC)7 | 142 | (AJ407071) |
- Listed are locus, species for which the locus was originally obtained and eretmodine species for which a specific locus was sequenced (for species designation see Rüber et al. 1999), microsatellite core sequence and size of cloned alleles, annealing temperature (Ta), citation for the primer sequences or eretmodine EMBL/GenBank accession numbers (this study), respectively. *For this study a new forward primer was designed: 5′-GCTGTG CTATTTAATCTGTTTATA-3′; †;primers reported in Zardoya et al. (1996) did not amplify in eretmodine cichlids and new internal forward 5′-AGCAGTGGAGCAGAGCGAGAACGC-3′ and reverse 5′-AATGTTCTGCGCCACTGGGTCTCC-3′ primers were designed based on the alignment given in Zardoya et al. (1996).
Microsatellite markers
Once suitable microsatellite loci were selected (see Results) amplified products were electrophoresed on 6% denaturating polyacrylamide gels using an ALF Express DNA sequencer (Pharmacia) with short glass plates. Gels were run between 2–4 h, depending on the allele size, at 55 °C (15 W, sampling interval 1 s). Alleles were sized with the program Allele LinksTM (Pharmacia) by using internal standards run in every lane. Internal size standards were obtained by PCR from M13mp18 + template DNA as described in van Oppen et al. (1997b). For locus TmoM25, a 453-bp sizer was designed for this study, using the reverse primer 5′-ATTTCGGAACCA CCATCAAA-3′ in combination with the universal M13mp18 + forward primer and an annealing temperature of 54 °C. The following size standards were used: 200 bp + 353 bp; 100 bp + 200 bp; 312 bp + 453 bp; 100 bp + 200 bp; 259 bp + 353 bp; and 100 bp + 200 bp for the loci TmoM5, TmoM11, TmoM25, Pzeb1, Pzeb3, and UNH002, respectively. Allelic corrections were performed for each locus using the sequenced alleles as references (Table 1) and, in addition, for each locus 2–3 randomly chosen individuals were run on all gels to ensure consistency of fragment sizing.
mtDNA
The proline tRNA with a segment of the mitochondrial control region was amplified using conditions and primers described in Rüber et al. (1999). Samples were sequenced using the AutoCyleTM Sequencing kit (Pharmacia) and run on an ALF express DNA sequencer (Pharmacia). Some samples were sequenced on an Applied Biosystem 373 A DNA sequencer using the Taq Dye Deoxy Terminator Cycle Sequencing Kit FS (PE Biosystems). The nucleotide sequence data were deposited in EMBL/GenBank under accession numbers AJ407050–AJ407077 for the sequenced microsatellite alleles and AJ407078–AJ407147 for the control region sequences.
Population genetic analyses
Genetic diversity based on microsatellite DNA and mtDNA was assessed for all populations. For the microsatellite data the genetic polymorphism was estimated for each population with genepop 3.1d (Raymond & Rousset 1995) as the number of alleles per locus (NA), the observed (HO) and the expected heterozygosity (HE). Samples were then tested for departures from Hardy–Weinberg equilibrium using probability tests with significance determined by the Markov chain method (Guo & Thompson 1992). Genotypic linkage disequilibrium was evaluated with genepop 3.1d for each pair of loci in each population and significance was determined through a log-likelihood based exact test (Goudet et al. 1996). mtDNA polymorphism was assessed by estimating both haplotype (H; Nei 1987) and nucleotide (π; Tajima 1983) diversity within populations. Differences in genetic diversity between the two species were assessed using the Mann–Whitney U-test (Sokal & Rohlf 1995).
Population structure
We tested for differences between populations in their allelic (microsatellites) or haplotypic (mtDNA sequences) distributions with an exact test of population differentiation (Raymond & Rousset 1995) as implemented in arlequin 2.000 (Schneider et al. 2000). The significance of the results was determined by the Markov chain method. For the microsatellite data, multilocus probabilities were produced using Fisher’s (1954) method of combining probabilities. We then quantified the extent of genetic differentiation among samples by calculating Weir & Cockerham’s (1984) estimators of F-statistics (θST) with arlequin 2.000. Significance of results was obtained through 5000 permutations. Table-wide rejection levels for multiple tests were calculated with sequential Bonferroni adjusted P-values throughout the analyses (Rice 1989).
We performed an analysis of molecular variance (amova), as described in Michalakis & Excoffier (1996), in order to partition the genetic diversity (based either on allele or haplotype frequency) into: (i) variance between species; (ii) variance among populations within species; and (iii) variance within populations. The significance of the variance components associated with the different levels of genetic structure was tested using nonparametric permutation procedures as implemented in arlequin 2.000.
For the microsatellite data, we assessed relationships among populations using chord distances (DCE; Cavalli-Sforza & Edwards 1967), computed for all pairwise comparisons with phylip version 3.572c (Felsenstein 1995). Pairwise distances were used to build a neighbour-joining (NJ) tree (Saitou & Nei 1987) and robustness of the tree topology was assessed through 1000 bootstrap replications (Felsenstein 1985). Phylogenetic relationships among mtDNA haplotypes were estimated using the NJ method based on Kimura 2-parameter corrected distances (Kimura 1980). Robustness of the inferred NJ tree was tested with paup* version 4.0b4a (Swofford 1997) using the bootstrap method with 1000 resamplings.
Finally, we explored the demographic history of the two species. To detect recent effective population size reductions from the microsatellite allele frequencies we used the program bottleneck 1.2.02 (Cornuet & Luikart 1996). This program uses the Sign and Wilcoxon tests to test for an excess of the observed heterozygosity which, for loci evolving under the infinite allele model (IAM), should be larger in populations having experienced a recent bottleneck than the heterozygosity expected for the observed number of alleles in populations under mutation-drift equilibrium. Another bottleneck signature tested with bottleneck 1.2.02 is a deviation of the allele frequency distribution from the approximately L-shaped distribution expected under mutation-drift equilibrium (Luikart et al. 1998). To test for a population expansion we applied Fu’s (1997)Fs-test of neutrality as implemented in arlequin 2.000, which is based on the comparison of rare and frequent mutations. After an episode of population growth, the coalescent theory predicts an excess of low frequency alleles and recent mutations in nonrecombining sequences compared to populations at equilibrium (Slatkin & Hudson 1991).
Results
Microsatellite DNA
Of 10 microsatellite loci tested six were chosen for population analyses on the basis of the repeat number in the sequenced alleles (Table 1). The selected loci showed at least 10 uninterrupted dinucleotide repeats in one of the individuals studied and were, therefore, considered to exhibit sufficient potential for polymorphism for population analyses. The six selected loci were: TmoM5, TmoM11, TmoM25, Pzeb1, Pzeb3, and UNH002.
The core sequences of four of the 10 microsatellite loci sequenced differed from those reported in the original studies. One out of two sequenced eretmodine specimens showed a perfect (AC)33 repeat compared to a compound-perfect (GC)n(AC)n repeat reported for locus TmoM5 in Tropheus moorii (Zardoya et al. 1996). In eretmodine cichlids locus TmoM25 had imperfect (CA)nCGT(CA)n alleles, compared to perfect (CA)n alleles found in T. moorii (Zardoya et al. 1996). The two eretmodine species sequenced for TmoM13 showed a perfect (CA)n repeat motif compared to a (CA)n(CG)n repeat motif found in T. moorii (Zardoya et al. 1996). The sequences available for locus Pzeb1 showed differences in the repeat in all species examined so far, and further indicate the presence of a poly T region following the microsatellite that also varies in length among alleles (Table 1). In eretmodines the repeat motives (GT)nGA(T)n and (GT)nTTGC(T)n were found.
Estimates of variability at the six microsatellite DNA loci within populations are shown in Table 2, and allele frequencies are presented in Appendix I. The total number of detected alleles per locus ranged from eight in Pzeb3 to 41 in TmoM5. The mean number of alleles (A) and expected heterozygosity (HE) across populations are significantly smaller (Mann–Whitney U-test; P = 0.05) in the Tanganicodus populations than in the Eretmodus populations (e.g. A, 4.7–5.7 compared to 8.5–13; HE, 0.41–0.49 compared to 0.74–0.82). At locus Pzeb3 all Tanganicodus populations are fixed for allele 313, which only occurs in a high frequency (> 0.500) in Eretmodus KOR. At locus UNH002 the Tanganicodus populations show either one or two alleles (160 and 162). Allele 160 is absent in the Eretmodus populations from KAM and KYE, and allele 162 is absent in Eretmodus from KOR (Appendix I).
| Eretmodus cyanosticus | Tanganicodus cf. irsacae | ||||||
|---|---|---|---|---|---|---|---|
| Locus | KAM (30) | KIK (34) | KOR (32)† | KYE (21) | KAM (25) | KAP (26) | KOR (18) |
| TmoM5 (41) | |||||||
| N A | 25 | 24 | 22 | 18 | 14 | 19 | 12 |
| H O | 1.00 | 0.94 | 0.91 | 0.90 | 0.72 | 1.00 | 0.89 |
| H E | 0.94 | 0.95 | 0.90 | 0.92 | 0.85 | 0.95 | 0.91 |
| Min (bp) | 274 | 274 | 272 | 268 | 284 | 286 | 282 |
| Max (bp) | 338 | 346 | 344 | 326 | 350 | 348 | 348 |
| Mode (bp) | 274 | 274 | 272 | 268 | 298 | 286 + 308 | 312 |
| TmoM11 (14) | |||||||
| N A | 6 | 7 | 7 | 3 | 6 | 2 | 3 |
| H O | 0.67 | 0.53 | 0.86 | 0.62 | 0.28*** | 0.04 | 0.17 |
| H E | 0.56 | 0.52 | 0.73 | 0.58 | 0.64 | 0.04 | 0.16 |
| Min (bp) | 153 | 153 | 147 | 159 | 153 | 167 | 165 |
| Max (bp) | 173 | 159 | 169 | 163 | 173 | 169 | 169 |
| Mode (bp) | 159 | 175 | 157 | 159 | 165 | 167 | 167 |
| TmoM25 (22) | |||||||
| N A | 14 | 17 | 14 | 9 | 7 | 8 | 6 |
| H O | 0.90 | 0.79 | 0.72** | 0.90 | 0.76 | 0.73 | 0.61 |
| H E | 0.88 | 0.89 | 0.93 | 0.86 | 0.67 | 0.69 | 0.60 |
| Min (bp) | 330 | 348 | 356 | 344 | 348 | 350 | 350 |
| Max (bp) | 392 | 386 | 382 | 368 | 378 | 374 | 374 |
| Mode (bp) | 352 | 354 | 364 | 354 | 356 | 350 | 350 |
| Pzeb1 (15) | |||||||
| N A | 11 | 10 | 2 | 9 | 5 | 3 | 4 |
| H O | 0.77 | 0.71 | 0.25 | 0.81 | 0.80 | 0.50 | 0.50 |
| H E | 0.82 | 0.81 | 0.38 | 0.79 | 0.79 | 0.62 | 0.53 |
| Min (bp) | 124 | 124 | 124 | 132 | 124 | 124 | 124 |
| Max (bp) | 146 | 164 | 126 | 156 | 144 | 138 | 140 |
| Mode (bp) | 124 | 124 | 126 | 140 | 138 | 124 | 124 |
| Pzeb3 (8) | |||||||
| N A | 6 | 8 | 5 | 4 | 1 | 1 | 1 |
| H O | 0.50*** | 0.38*** | 0.53* | 0.48 | 0.00 | 0.00 | 0.00 |
| H E | 0.82 | 0.73 | 0.58 | 0.65 | 0.00 | 0.00 | 0.00 |
| Min (bp) | 313 | 313 | 311 | 315 | 313 | 313 | 313 |
| Max (bp) | 325 | 329 | 321 | 325 | 313 | 313 | 313 |
| Mode (bp) | 321 | 319 | 313 | 325 | 313 | 313 | 313 |
| UNH002 (22) | |||||||
| N A | 16 | 12 | 16 | 8 | 1 | 2 | 2 |
| H O | 0.90 | 0.82 | 0.97 | 0.57* | 0.00 | 0.15 | 0.39 |
| H E | 0.91 | 0.85 | 0.91 | 0.67 | 0.00 | 0.14 | 0.32 |
| Min (bp) | 154 | 158 | 160 | 162 | 160 | 160 | 160 |
| Max (bp) | 200 | 188 | 214 | 188 | 160 | 162 | 162 |
| Mode (bp) | 178 | 168 | 170 | 162 | 160 | 160 | 160 |
| TOTAL | |||||||
| (A) | 13.0 ± 7.16 | 13.0 ± 6.45 | 11.0 ± 7.59 | 8.50 ± 5.32 | 5.70 ± 4.80 | 5.80 ± 6.91 | 4.70 ± 3.98 |
| (H0) | 0.79 ± 0.18 | 0.70 ± 0.21 | 0.71 ± 0.27 | 0.71 ± 0.18 | 0.43 ± 0.38 | 0.40 ± 0.41 | 0.43 ± 0.32 |
| (HE) | 0.82 ± 0.14 | 0.79 ± 0.15 | 0.74 ± 0.22 | 0.75 ± 0.13 | 0.49 ± 0.39 | 0.41 ± 0.40 | 0.42 ± 0.33 |
- Sample size is indicated in parentheses beneath each population. The total number of alleles scored for each locus is indicated in parentheses next to the locus name. NA, number of alleles; HO, observed heterozygosity; HE, expected heterozygosity. Allelic diversity (A), average observed (HO), and expected Hardy–Weinberg (HE) heterozygosity with standard deviation for each population. Size range of alleles: Min (bp), Max (bp), and Mode (bp) are provided for each population and locus. †Three E. cyanostictus individuals from KOR had one allele of size 127 at locus TmoM11, which is 10 bp shorter than the entire microsatellite flanking region, suggesting deletion in the flanking region. For these individuals, genotypes were coded as missing. Significant deviations from Hardy–Weinberg equilibrium: *P < 0.05; **P < 0.01; ***P < 0.001.
Six of 42 single-locus tests for deviation from Hardy–Weinberg equilibrium gave significant results, but only three [TmoM11 in Tanganicodus (KAM) and Pzeb3 in Eretmodus (KAM and KIK)] remained significant when adjusted for table-wide significance by a sequential Bonferroni procedure. Linkage disequilibrium was observed in three out of 105 pairwise comparisons [E. cyanostictus KAM, TmoM5 and TmoM25 (P = 0.050), TmoM25 and Pzeb1 (P = 0.015); E. cyanostictus KYE, TmoM25 and Pzeb3 (P = 0.007)] but none of them remained significant after sequential Bonferroni correction.
mtDNA
The 198 DNA sequences comprised 70 mitochondrial haplotypes (Table 3 and Appendix II). The overall haplotype and nucleotide diversity were 0.947 and 0.040, respectively. Haplotype diversity was significantly smaller (Mann–Whitney U-test; P = 0.05) for the Tanganicodus populations than for the Eretmodus populations but not so for the nucleotide diversity (see Discussion). For these comparisons, two populations (Eretmodus KAP and Tanganicodus KIK) were left out due to small sample sizes. Two haplotypes (37 and 66) were shared among individuals classified as Eretmodus and Tanganicodus (Appendix II).
| N | NH | S | H | π | |
|---|---|---|---|---|---|
| Eretmodus | |||||
| KAM | 30 | 15 | 18 | 0.878 ± 0.045 | 0.0066 ± 0.004 |
| KIK | 34 | 24 | 26 | 0.975 ± 0.014 | 0.0112 ± 0.006 |
| KAP | 6 | 4 | 4 | 0.800 ± 0.174 | 0.0071 ± 0.005 |
| KOR | 32 | 10 | 9 | 0.853 ± 0.040 | 0.0062 ± 0.004 |
| KYE | 21 | 12 | 10 | 0.905 ± 0.048 | 0.0064 ± 0.004 |
| Tanganicodus | |||||
| KAM | 25 | 4 | 18 | 0.417 ± 0.115 | 0.0127 ± 0.007 |
| KIK | 6 | 4 | 24 | 0.867 ± 0.129 | 0.0369 ± 0.022 |
| KAP | 26 | 4 | 7 | 0.286 ± 0.112 | 0.0042 ± 0.003 |
| KOR | 18 | 3 | 7 | 0.582 ± 0.061 | 0.0035 ± 0.003 |
| TOTAL | 198 | 70 | 63 | 0.947 ± 0.009 | 0.0404 ± 0.020 |
- Listed are: N, number of individuals; NH, number of haplotypes; S, number of segregating sites; H, average haplotype diversity ± SD, average nucleotide diversity ± SD.
| E. cyanostictus | T. cf. irsacae | 1111111111111111111111111112222222222222222233333 14477788888991122223333344455566666777780011124445557899900223 110914801234131723572356828927913478157812346705782466734737894 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype | KAM 30 | KIK 34 | KAP 6 | KOR 32 | KYE 21 | KAM 25 | KIK 6 | KAP 26 | KOR 18 | |
| 1 | 9 | 4 | GTCATTTCAAGAATCATCATGGTAAAAAATAACATTAATTTTCGTATTGTTTCGGTTTCAAGT | |||||||
| 2 | 6 | ...........................................C................... | ||||||||
| 3 | 2 | A.........................................................T.... | ||||||||
| 4 | 2 | 1 | .......T...................................C....A.............. | |||||||
| 5 | 1 | ..........................................................T.... | ||||||||
| 6 | 1 | ................C..........................C................... | ||||||||
| 7 | 1 | ......................C..........................C............. | ||||||||
| 8 | 1 | .......................T...................C................... | ||||||||
| 9 | 1 | ........G...................................................... | ||||||||
| 10 | 1 | .......... G..............................................T.... | ||||||||
| 11 | 1 | ......................C........................................ | ||||||||
| 12 | 1 | ...........................................T....A.............C | ||||||||
| 13 | 1 | ............................................................GTC | ||||||||
| 14 | 1 | ............................G..............C......C............ | ||||||||
| 15 | 1 | .............C...............A.............T................... | ||||||||
| 16 | 3 | ...................C...........G............................... | ||||||||
| 17 | 2 | .....C..........................................A......A....... | ||||||||
| 18 | 2 | ...........................................A...............G... | ||||||||
| 19 | 2 | ......................................................A........ | ||||||||
| 20 | 2 | ....................................................T.......... | ||||||||
| 21 | 2 | ..............T....C...........G................A.............. | ||||||||
| 22 | 1 | ...........................................TC.................. | ||||||||
| 23 | 1 | ..............T....C........................................... | ||||||||
| 24 | 1 | ...................................C........................... | ||||||||
| 25 | 1 | ...........................................AC.................. | ||||||||
| 26 | 1 | ...................C...........G...........A..C.A.............. | ||||||||
| 27 | 1 | ...........................................AC..............G... | ||||||||
| 28 | 1 | ..............T....C..........C..G...G.....C...............G..C | ||||||||
| 29 | 1 | A..........................................TC.................. | ||||||||
| 30 | 1 | ............................................CG....C............ | ||||||||
| 31 | 1 | ............................................C.................. | ||||||||
| 32 | 1 | ...................C...........G..C............................ | ||||||||
| 33 | 1 | ...................C...........G...........................G... | ||||||||
| 34 | 1 | ...............G.........................C..................... | ||||||||
| 35 | 1 | ...........................................T................... | ||||||||
| 36 | 1 | ..........................G..........................A......... | ||||||||
| 37 | 1 | 1 | ...................C..........................C................ | |||||||
| 38 | 2 | ..............T....C..........C..G..G....C.C....A..........G..C | ||||||||
| 39 | 3 | ...........................................C................GT. | ||||||||
| 40 | 1 | .C..........................G..............C......C............ | ||||||||
| 41 | 6 | ......CT...........CAACG......C.......CC...C...C.........CTG.CC | ||||||||
| 42 | 3 | ......CT...G.......CAAC.......C.......CC...C...C.........CTG.CC | ||||||||
| 43 | 2 | ......CT...G.......CAACG......C.......CC...C...C.........CTG.CC | ||||||||
| 44 | 2 | ......CT...........CAAC.......C.......CC...C...C.........CTG.CC | ||||||||
| 45 | 1 | ......CT...........CAAC.......C.......CC...C...CA........CTG.CC | ||||||||
| 46 | 1 | ......CT...G.......CAACG......C.......CC...C...CA........CTG.CC | ||||||||
| 47 | 1 | ......CT...GG......CAAC.......C.......CC...C...C.........CTG.CC | ||||||||
| 48 | 1 | ......CT...........CAAC.......C.......CC..TC...CA........CTG.CC | ||||||||
| 49 | 1 | ......CT...........CAACG.G....C.......CC...C...CA.C......CTG.CC | ||||||||
| 50 | 1 | ......CT...........CAACG......C.......CC.C.C...C.........CTG.CC | ||||||||
| 51 | 1 | ......CT...........CAAC.G.....C.......CC...C...CA.C......CTG.CC | ||||||||
| 52 | 1 | ......CT...........CAACG......C.......CC...C...CA....A...CTG.CC | ||||||||
| 53 | 19 | .CTC....G.A......TCC..........C...C........C.....A......C.TGGT. | ||||||||
| 54 | 2 | .CTC......A......TCC..........C...C........C.....A......C.TGGT. | ||||||||
| 55 | 2 | .CTC......A...T. TCC..........C.T.C........C.....A......C.TGGTC | ||||||||
| 56 | 1 | .CTC....G.A......TCC..........C...C........C.....A......C.TGGTC | ||||||||
| 57 | 3 | 10 | ..T.......A......TCC.......G.CC.T.C........C....AA.C....C.TGGTC | |||||||
| 58 | 5 | ..T.......A......TCC.........CC.T.C........C....AA.C....C.TGGTC | ||||||||
| 59 | 1 | 5 | ..T.....G.A......TCC.......G.CC.T..... CC..C....AA.C....C.TGGTC | |||||||
| 60 | 2 | ..T.......A......TCC.......G.CC.T.C........C....AA.C....CCTGGTC | ||||||||
| 61 | 1 | 2 | ..T.....G.A......TCC.......G.CC.T.C........C....AA.C....C.TG.TC | |||||||
| 62 | 1 | ..T.......A......TCC.......G.CC.T.C........C…...AA......C.TGGTC | ||||||||
| 63 | 1 | ..T.......A......TCC.......G.CC.T.C....C...C....AA.C....C.TGGTC | ||||||||
| 64 | 1 | ..T.......A......TCC.......G.CC.T.C....C........AA.A....C.TGGTC | ||||||||
| 65 | 1 | 1 | ..T..............TCC.......G.CC.T.C.....C..C....AA.C....C.TGGTC | |||||||
| 66 | 4 | 22 | 9 | ..T.......A......TCC.......G.CC.T.C.....C..C....AA.C....C.TGGTC | ||||||
| 67 | 8 | ..T......TA......TCC.......G.CC.T.C.....C..C....AA.C....C.TGGTC | ||||||||
| 68 | 2 | 1 | ..TC......A......TCC.......G..C...C........C....AA.A....C.TG.TC | |||||||
| 69 | 1 | ..T.......A......TCC.......G.CC.T.C.....C..C....AA......C.TGGTC | ||||||||
| 70 | 1 | ..TCC.....A......TCC.......G..C...C........C....AA.A....C.TG.TC | ||||||||
- Haplotypes 1–52 clustered in Lineage C whereas haplotypes 53–70 clustered in Lineage D (see Fig. 1).
Population structure
Across all populations, differentiation tests based on microsatellites showed highly significant heterogeneity of allele frequencies (P << 0.001). Single locus pairwise tests were all significant except for eight comparisons [E. cyanostictus KAM and KIK (TmoM11, TmoM25, Pzeb1); E. cyanostictus KIK and KYE (TmoM25); T. cf. irsacae KOR and KAP (TmoM5, TmoM25, UNH002); T. cf. irsacae KAM and KAP (UNH002)]. Multilocus probabilities of heterogeneity between population pairs were all highly significant (P < 0.001; for T. cf. irsacae KOR and KAP, P < 0.01).
Based on the microsatellite data, estimates of overall θST indicated significant levels of genetic variance among all populations (θST = 0.250, P << 0.001). For the four Eretmodus populations the value was θST = 0.127 (P << 0.001) and for the three Tanganicodus populations it was θST = 0.240 (P << 0.001). All population pairwise estimates of multilocus θST were significantly different from zero (Table 4). Our estimates of fixation indices are not substantially affected and remain significantly different from zero when loci TmoM11 and Pzeb3, which showed significant deviation from Hardy–Weinberg equilibrium in a few populations (see Table 2), are excluded.
| E. cyanostictus | T. cf. irsacae | ||||||
|---|---|---|---|---|---|---|---|
| KAM | KIK | KOR | KYE | KAM | KAP | KOR | |
| KAM | 0.024** | 0.138** | 0.094** | 0.294** | 0.338** | 0.313** | |
| KIK | 0.037 * | 0.162** | 0.121** | 0.299** | 0.346** | 0.323** | |
| KOR | 0.135 ** | 0.086 ** | 0.227** | 0.266** | 0.272** | 0.290** | |
| KYE | 0.109 ** | 0.059 ** | 0.122 ** | 0.382** | 0.365** | 0.410** | |
| KAM | 0.344 ** | 0.289 ** | 0.354 ** | 0.348 ** | 0.295** | 0.297** | |
| KAP | 0.408 ** | 0.351 ** | 0.350 ** | 0.421 ** | 0.649 ** | 0.040* | |
| KOR | 0.258 ** | 0.205 ** | 0.223 ** | 0.252 ** | 0.509 ** | 0.258 ** | |
- Significance of FST-values: *P < 0.01; **P < 0.001.
Population differentiation tests based on mtDNA haplotype frequencies were highly significant for all pairwise comparisons (P << 0.001; E. cyanostictus KAM and KIK, P = 0.0015). Overall, estimates of θST indicated significant levels of genetic variance among all populations (θST = 0.263, P << 0.001). For the four Eretmodus populations the value was θST = 0.089 (P << 0.001) and for the three Tanganicodus populations it was θST = 0.526 (P << 0.001). Population pairwise estimates of θST were all significantly different from zero (Table 4).
The amova revealed that 18.4% of the total variance in microsatellite allele frequency was due to between species heterogeneity (FCT = 0.184) and 12.9% was caused by differences among populations within species (FSC = 0.158). The remainder (68.7%) was due to variation within populations (FST = 0.312). For the mtDNA data, the corresponding values were 7.8% (FCT = 0.078), 21.1% (FSC = 0.228) and 71.1% (FST = 0.289, Table 5).
| Variance component Source of variation | V | % | Fixation indices | P |
|---|---|---|---|---|
| Microsatellites | ||||
| Between species | 0.52 | 18.37 | F CT 0.184 | < 0.0000 |
| Among populations/ within species | 0.37 | 12.88 | F SC 0.158 | < 0.0000 |
| Within populations | 1.96 | 68.75 | F ST 0.312 | < 0.0000 |
| mtDNA sequences | ||||
| Between species | 0.04 | 7.80 | F CT 0.078 | < 0.0000 |
| Among populations/ within species | 0.11 | 21.09 | F SC 0.228 | < 0.0000 |
| Within populations | 0.36 | 71.11 | F ST 0.289 | 0.0508 |
Population clustering and phylogenetic analysis
A majority-rule consensus NJ tree (based on 1000 bootstrap-replicated chord distance matrices) grouped most populations by species rather than geographical location (Fig. 2). All except one internal node were well supported (i.e. bootstrap values > 70%). For example, the node grouping all Eretmodus was supported by a bootstrap value of 91% (Fig. 2).

Unrooted, majority-rule consensus tree representing 1000 bootstrap replicates of chord distance data (DCE), cluster with the neighbour-joining algorithm. Bootstrap values based on 1000 replications are shown above branches.
The NJ phylogeny of the 70 identified mtDNA haplotypes shows a basal split into two genetically distinct lineages (bootstrap values 94% and 86%; Fig. 1). These lineages were previously defined as the E. cyanostictus lineage C and the T. cf. irsacae lineage D (Rüber et al. 1999). Within lineage C two major clades were observed, one comprising all the E. cyanostictus from KYE (clade C-1, bootstrap value 100%) and another that includes all individuals of E. cyanostictus from the localities KAM and KIK (clade C-2, bootstrap value 75%). Clade C-2 also includes a haplotype shared by one individual of E. cyanostictus from KIK and one of T. cf. irsacae from KIK (37), and three haplotypes found in a few individuals of T. cf. irsacae from either KAM or KIK (38–40, Appendix II).
Within lineage D three major clades were found (bootstrap values 62%, 68% and 97%). Clade D-3 comprises four haplotypes (53–56) representing 24 individuals of T. cf. irsacae from KAM and KIK. Clade D-2 containes haplotypes 68 and 70 from three T. cf. irsacae (localities KAP and KOR). And finally, clade D-1 contains 12 haplotypes. In this clade, haplotypes 57–65 are found in E. cyanostictus from KAP and KOR; haplotype 66 is found in four E. cyanostictus from KOR, in nine T. cf. irsacae from KOR, and in 22 T. cf. irsacae from KAP; the remaining two haplotypes (67 and 69) are found in eight T. cf. irsacae from KOR and in one T. cf. irsacae from KAP. The average sequence divergence between individuals from lineage C and D was 6.20% ± 0.12% (net sequence divergence corrected for intrapopulation polymorphism was 4.45% ± 0.15%). Within lineage C and D the average sequence divergence between individuals was 2.56% ± 0.10 and 1.36% ± 0.03, respectively. Applying a molecular clock calibration based on cichlid control region sequences (5.6% per site per 106 years; Nagl et al. 2000) on the net sequence divergence suggests that the two lineages (C and D) separated roughly 0.8 million years ago.
Demographic history
Both the Sign and the Wilcoxon tests gave no indication for heterozygote excess, and the allele frequency distributions indicated no deviation from an approximate L-shaped distribution in any of the populations. Thus, these results indicate that, according to the microsatellite data, none of the observed populations underwent a recent bottleneck. Fu’s test supported a demographic expansion of the Eretmodus populations with the exception of Eretmodus KOR (FS-values ranged from −6.91 to −13.26; P ≤ 0.0001; Eretmodus KOR FS = −2.79, P = 0.0718. All Tanganicodus populations showed positive FS-values and are therefore compatible with population stasis (FS-values ranged from 1.16 to 5.54; P > 0.750).
Discussion
Microsatellite sequences
In eretmodine cichlids four out of 10 loci have undergone mutational events in the microsatellite repeats, compared to the species from which the microsatellites were obtained (Table 1). Differences in the repeat motif as found here underscore the need for more DNA sequence data to adequately assess genetic variability within and among species, and to understand the evolutionary processes of these markers (e.g. Zardoya et al. 1996). Such information may also prove essential to correct for size-homoplasies in estimating demographic population parameters such as structure, gene flow, and effective population size (e.g. Angers & Bernatchez 1997). Another illustration of this issue was given in a recent study that revealed extensive size homoplasy at locus Pzeb4 within and among 11 Lake Malawi cichlids species (van Oppen et al. 2000). These authors further cautioned against using population parameters based on the stepwise mutation model (SMM) if conformance to SMM for the loci used were not demonstrated. Our results indicate that loci TmoM11, Pzeb3, and UNH002, which exhibit perfect dinucleotide repeats in all the cichlid species studied so far, might be less affected by size homoplasy than other loci that show more complex mutations at the intra- and interspecific level (Table 1). Further comparative data of other cichlid species might help to distinguish loci that are more affected by size homoplasy than others.
Intra- and interspecific genetic variation
Genetic variation inferred from mtDNA sequences and microsatellites was consistently lower in the Tanganicodus than in the Eretmodus populations (Tables 2 and 3). Only the nucleotide diversity did not differ significantly between the two species due to haplotypes 39 and 40 found in T. cf. irsacae at KAM (Table 3) that clustered with Eretmodus haplotypes in clade C-2 (Fig. 1). Excluding these haplotypes from the analysis reduces the nucleotide diversity of the Tanganicodus KAM population to 0.0005 ± 0.001 resulting in a significantly lower nucleotide diversity in all the Tanganicodus populations in comparison to the Eretmodus populations (Mann–Whitney U-test; P = 0.05). The relatively high nucleotide diversity observed in Tanganicodus KIK, which was not included in the analyses (see Results), can also be explained by the presence of two haplotypes (37 and 38, found in three individuals) that clustered with clade C-2 (Fig. 1).
The observed species-specific differences in genetic diversity suggest that the two species might have had different evolutionary histories in terms of random genetic drift, variation of effective population size, or natural selection. We estimated NE for the Eretmodus and Tanganicodus populations using the formula π = 4NEµ, where π is the nucleotide diversity and µ is the mutation rate assumed to be 2.8 × 10−8 (substitutions × base pair−1 × year−1) for the control region in cichlids (Nagl et al. 2000). For the Eretmodus populations the estimated NE is in the order of 2–4 × 105, whereas for the Tanganicodus populations it is in the order of 2–10 × 104. Parker & Kornfield (1997) and Nagl et al. (1998) obtained estimates of either Nf or NE of similar magnitude for several species of lakes Malawi and Victoria cichlids. The difference in the estimated effective population size between the two eretmodine species agrees with field observations by Hori et al. (1983), who studied the abundance and microdistribution of cichlids on a rocky shore in the northern part of the lake and recorded higher abundance of E. cf. cyanostictus (lineage A) than of T. irsacae (lineage A).
The mtDNA control region sequences revealed a relatively large amount of polymorphism in the Eretmodus populations in comparison to other studies on lacustrine cichlids. For example, Meyer et al. (1996) found an overall nucleotide diversity in the mtDNA control region of 0.0026 and 0.0037 in two Simochromis species (25 and 28 individuals studied) from Lake Tanganyika collected over a shoreline of 440 km. Bowers et al. (1994) found an overall nucleotide diversity of 0.0008 and 0.0050 in two species of the mbuna genus Melanochromis from the southern part of Lake Malawi (68 and 42 individuals studied). They also found that 75% of the populations were fixed for a single haplotype. A collection of all available Lake Malawi mbuna control region sequences revealed 58 control region haplotypes with a total of 46 segregating sites among 180 individuals from 34 species representing several mbuna genera [Parker & Kornfield (1997); see Table 3 for a comparison with the two species studied in this paper]. The low levels of mtDNA variation found in many Lake Malawi mbunas may be due to recent colonization of habitat patches from refugia populations in the recent past due to lake level fluctuations (van Oppen et al. 1997a; Arnegard et al. 1999; Markert et al. 1999; Sturmbauer et al. 2001). The level of expected heterozygosity based on microsatellite data found in the Eretmodus populations (0.72 average across all loci and all populations), is comparable with data for Melanochromis auratus (0.67; Markert et al. 1999), Labeotropheus fulleborni (0.83; Arnegard et al. 1999) and for four Pseudotropheus species (0.72 average over species; van Oppen et al. 1997a) from Lake Malawi. In the light of the findings from this and other studies, T. cf. irsacae is the genetically least polymorphic lacustrine cichlid species found so far. There is no evidence for a recent population bottleneck that may account for the lower genetic variation found in the Tanganicodus populations. On the other hand, we found evidence of a population expansion in Eretmodus, the species that shows higher genetic diversity. With the information at hand it is not possible to determine whether the higher genetic diversity in Eretmodus is only associated with the hypothesized expansion or if there are additional species-specific differences in life history traits and/or environmental factors that might affect their population sizes.
Population structure
Both microsatellite markers and mtDNA sequences revealed significant levels of genetic structure within the two sympatric eretmodine species — along 138 km of coastline for the Eretmodus populations and over a distance of 35 km for the Tanganicodus populations. The higher degree of population structuring observed in Tanganicodus may be attributed to the lower levels of genetic diversity, in both microsatellites and mtDNA markers within populations, that influence measures of population differentiation (e.g. Nagylaki 1998). Other factors such as dispersal ability and phylopatry may contribute to differences in population structure between the two species.
Among the 70 identified haplotypes, shared haplotypes were never found between more than two neighbouring populations. For example the Eretmodus populations at KAM and KIK with 15 and 24 haplotypes, respectively, separated by a distance of 7 km of rocky shore, shared only two haplotypes. In the Tanganicodus populations (Appendix II) haplotype 67 was observed in over 40% of the individuals from KOR, but not at KAP which is situated only 7 km away. Such a low frequency of shared mtDNA haplotypes among localities is likely to be an indicator for the expressed site fidelity of eretmodine cichlids, which may rapidly lead to genetic isolation among populations. Although the amount of shared mtDNA haplotypes may change with increasing sample size, our microsatellite results support a high degree of genetic isolation between populations.
Philopatry coupled with the patchy distribution of rocky habitats along the lake shores have been identified as important factors for population divergence among Lake Malawi mbuna cichlids (van Oppen et al. 1997a; Arnegard et al. 1999; Markert et al. 1999). The availability of suitable habitat patches as well as their distribution is strongly affected by lake level fluctuations. A recent study confirmed that lake level fluctuations in Lake Tanganyika, induced by tectonic and/or climatic events, have strongly affected the distribution and fragmentation of rocky habitat patches in the littoral (Cohen et al. 1997). Cohen et al. (1997) identified seven lake lowstand periods in the Northern Tanganyika basin that have been named and dated as follows: (f) 1.1 Ma; (d) 393–363 Ka (1 Ka = 1000 years); (c) 295–262 Ka; (b) 193–169 Ka; (a) 40–35 Ka; (x) 23 Ka; (y) 18 Ka. The rise of the lake to its present level was completed 12 Ka (A. Cohen, personal communication; see also Sturmbauer et al. 2001). The magnitudes of the lake lowstands below present lake level were given by these authors as 650–700 m for (f); 350 m for (d); 350 m for (c), 250 m for (b) and 160 m for (a). Gasse et al. (1989) give estimates of 300–400 m for the magnitude of the late Pleistocene lowstands (x) and (y). Other authors suggest them to be more likely in the order of 150–200 m (A. Cohen, personal communication). Lake level fluctuations in the Holocene were less severe in Lake Tanganyika (maximum 75 m; see Coulter 1991) compared to Lake Malawi where it has been reported that a lake level drop of 120 m occurred 200 years ago (Owen et al. 1990).
As they may cause fusion or isolation of populations, water level changes in shallow areas of the lake are likely to have severe effects on population structure. At steeply sloping shores, lake level fluctuations will mostly cause a vertical dislocation of the populations and, as in these zones rocky habitats often extend far into deep water, are less likely to promote cycles of fusions and isolations of populations (Sturmbauer et al. 1997; Sturmbauer 1998). This is the case for all localities analysed in this study. We provide evidence for significant population differentiation along a mostly continuous rocky shoreline that is only rarely interrupted by few small sandy bays and influent rivers, with areas of sand deposition that might act as barriers to gene flow, and that may not have been much affected by lake level fluctuations. A question that needs to be studied in more detail is whether the small sandy beaches observed in the study area act as effective dispersal barriers and significantly contribute to population differentiation in the rock-dwelling eretmodine cichlids, or whether divergence also occurs under parapatric conditions —, i.e. along a continuous habitat (Taylor et al. 2001).
Evidence for introgression
Throughout its narrow distribution range T. cf. irsacae occurs in sympatry with E. cyanostictus. Chord distances grouped the Eretmodus populations in a separate cluster (Fig. 2). The mitochondrial gene phylogeny (Fig. 1), on the other hand, shows that some individuals of T. cf. irsacae from KAM and KIK were placed within the E. cyanostictus lineage (clade C-2), whereas all individuals of E. cyanostictus from KAP and KOR were resolved within the T. cf. irsacae lineage (clade D-1).
The observed distribution of mitochondrial haplotypes in these two closely related species is incongruent with the morphology-based species assignment which clearly indicates their morphological distinctness. Shared ancient polymorphism as a result of incomplete lineage sorting or introgression during secondary contact may be responsible for this incongruence. At the present state of knowledge we favour the introgression scenario over incomplete lineage sorting. An argument against incomplete lineage sorting is the local occurrence of the mtDNA haplotypes found in lineages C and D that conflict with the species tree. In addition, given the average sequence divergence of 6.20% separating the two lineages, we consider it unlikely that retained ancestral haplotypes in Eretmodus at KOR and KAP would be identical or nearly identical to the Tanganicodus haplotypes at those localities. There are five fixed substitutions between lineage C and D haplotypes. Lineage D haplotypes show the following diagnostic substitutions: 40T, 123 T, 125 C, 248 A, and 294 C. Two of these sites show transversions (125 A/C and 248 T/A) which are diagnostic for lineage D haplotypes in regard to all other eretmodine lineages [based on a total of 288 eretmodine control region sequences; this study and Rüber et al. (1999)]. Therefore, we consider it more likely that the unique transversional mutations at sites 125 and 248 occurred along the branch grouping all lineage D haplotypes rather than it being present as a polymorphism in the shared ancestral gene pool from which E. cyanostictus, T. cf. irsacae and other eretmodine cichlids may have originated.
The possible mtDNA introgression cases we observe in our data are spatially restricted and asymmetric: complete introgression from Tanganicodus to Eretmodus at KOR and KAP, and partial introgression from Eretmodus to Tanganicodus at KIK and KAM. In contrast, the microsatellite allele frequency distributions indicate a limited degree of nuclear introgression, although allele 313 at locus Pzeb3 which is fixed in the Tanganicodus populations appears in a relatively high frequency in Eretmodus at KOR (see Results and Appendix I). Moreover, introgressed populations do not differ phenotypically from nonintrogressed ones. These observations, together with the genetical differentiation observed between sympatric Tanganicodus and Eretmodus populations (Table 4), suggests that hybridization might have taken place locally for a short period of time during secondary contact, and that there is no ongoing gene flow. Reinforcement may have contributed to the rapid evolution of reproductive isolation of the two sister species upon secondary contact, despite continuing sympatry (Butlin 1989).
The fixation of Tanganicodus mtDNA in the introgressed Eretmodus populations at KAP and KOR may have occurred either by chance (via founder effects or drift) or by selection. Although we have no indication that selection is responsible for the fixation of Tanganicodus mtDNA in the two introgressed Eretmodus populations, selective advantages of heterospecific mtDNA have been suggested as explanations of similar cases among salmonid species (Wilson & Bernatchez 1998 and references therein). The proposed direction of mtDNA introgression from Tanganicodus to Eretmodus at KAP and KOR seems at odds with the observation of higher genetic diversity and hence larger population sizes found in the latter populations. It is possible, however, that small Eretmodus founder populations colonized this area of the lake and established secondary contact with Tanganicodus populations, and that the presumed expansion of population size in Eretmodus happened after the interspecific replacement of mtDNA due to introgression.
There is increasing evidence, particularly from fishes, that mtDNA introgression between allopatrically diverged taxa in zones of secondary intergradation may be more common than previously thought (e.g. Dowling & deMarais 1993; Bernatchez et al. 1995; Wilson & Bernatchez 1998). However, it remains to be tested to what extent historic events of introgressive hybridization can explain patterns of shared haplotypes among species, or even genera, in other cichlid species flocks. Incomplete lineage sorting has been postulated for the lakes Malawi and Victoria cichlid flocks (Moran & Kornfield 1993, 1995; Parker & Kornfield 1997; Nagl et al. 1998, 2000). As in Lake Tanganyika, intermediate phenotypes are rare or absent in these lakes, suggesting that little or no hybridization is going on at the present time, possibly due to strong assortative mating (Seehausen et al. 1997; Knight et al. 1998; van Oppen et al. 1998). In contrast to Lake Tanganyika, the ages of these radiations are much younger, which makes incomplete lineage sorting more likely. Nevertheless, introgression caused by, for example, changes in water transparency, which may have occasionally broken down reproductive barriers among multiple species (see Seehausen et al. 1997), may in some cases be an alternative hypothesis.
Acknowledgements
We are grateful to Heinz H. Büscher for inviting L.R. to a collection trip to the Democratic Republic of Congo in 1996. Thanks to Rafael Zardoya for his help with the cloning of some of the microsatellites; Paul Shaw and Peter Gallbusera for primer exchange and sharing protocols. L.R. also thanks Sanja Baric, Harald Niederstätter and Walter Salzburger for their help and Hospitality at the Zoological Institute, University of Innsbruck, where parts of the microsatellite cloning and mtDNA sequencing was conducted. We gratefully acknowledge the help of T. Backeljau, K. Martens and S. Villalba for reading and commenting on early drafts of the manuscript and/or discussions. We also thank three anonymous reviewers for useful suggestions and comments on an earlier version of this manuscript. D. Swofford kindly granted permission to publish results based on the test version of paup* 4.0d65. This research was supported by grants from the Swiss National Science Foundation (TMR 83EU-045301), and the Gottfried R. Friedli-Stiftung (Bülach, Switzerland) to L.R., the Roche Research Foundation (Basel, Switzerland) to L.R. and A.M. Additional support came from a DWTC project to E.V., US NSF grants BSR-9107838 and DEB-9615178 and support from the University of Konstanz to A.M and grant P12339-GEN from the Austrian research Foundation to C.S.
Appendix
| Eretmodus cyanostictus | Tanganicodus cf. irsacae | ||||||
|---|---|---|---|---|---|---|---|
| Allele length (bp) | KAM | KIK | KOR | KYE | KAM | KAP | KOR |
| TmoM5 | |||||||
| 268 | — | — | — | 0.190 | — | — | — |
| 272 | — | — | 0.281 | — | — | — | — |
| 274 | 0.200 | 0.162 | 0.109 | 0.048 | — | — | — |
| 276 | 0.033 | 0.074 | 0.063 | — | — | — | — |
| 278 | — | — | 0.016 | — | — | — | — |
| 280 | 0.017 | — | — | — | — | — | — |
| 282 | 0.017 | — | — | — | — | — | 0.028 |
| 284 | — | 0.088 | — | — | 0.240 | — | — |
| 286 | 0.067 | — | — | — | 0.020 | 0.135 | 0.111 |
| 288 | 0.050 | — | — | — | — | — | — |
| 290 | — | — | — | 0.024 | — | — | — |
| 292 | 0.033 | 0.015 | 0.031 | 0.048 | — | — | 0.056 |
| 294 | 0.033 | 0.044 | 0.031 | — | 0.040 | 0.038 | — |
| 296 | 0.100 | 0.044 | 0.016 | 0.024 | 0.120 | — | — |
| 298 | 0.017 | 0.015 | — | — | 0.280 | — | — |
| 300 | — | 0.059 | 0.078 | 0.024 | 0.040 | 0.019 | — |
| 302 | — | 0.074 | 0.031 | 0.048 | — | 0.058 | — |
| 304 | 0.033 | 0.015 | 0.031 | 0.024 | — | — | 0.028 |
| 306 | 0.017 | 0.029 | 0.031 | 0.048 | 0.060 | 0.058 | — |
| 308 | 0.067 | 0.059 | 0.031 | 0.024 | 0.020 | 0.135 | 0.111 |
| 310 | 0.050 | 0.015 | — | 0.024 | — | 0.058 | 0.111 |
| 312 | 0.033 | — | 0.016 | 0.071 | — | 0.077 | 0.194 |
| 314 | 0.033 | 0.059 | 0.016 | 0.143 | — | 0.038 | 0.111 |
| 316 | — | 0.015 | — | 0.143 | — | 0.058 | 0.139 |
| 318 | 0.017 | 0.044 | 0.047 | 0.024 | — | 0.038 | 0.028 |
| 320 | 0.050 | — | — | 0.024 | — | 0.019 | — |
| 322 | — | — | 0.078 | 0.024 | — | 0.038 | — |
| 324 | 0.017 | 0.044 | — | — | 0.020 | — | — |
| 326 | 0.017 | 0.029 | 0.016 | 0.048 | — | — | — |
| 328 | 0.017 | 0.029 | 0.016 | — | — | — | — |
| 330 | 0.033 | — | 0.016 | — | — | — | — |
| 332 | 0.017 | — | — | — | 0.020 | 0.058 | 0.056 |
| 334 | 0.017 | 0.015 | 0.016 | — | 0.020 | 0.058 | — |
| 336 | — | 0.015 | 0.016 | — | — | 0.038 | — |
| 338 | 0.017 | 0.015 | — | — | 0.040 | 0.038 | — |
| 340 | — | — | — | — | — | 0.019 | — |
| 342 | — | 0.015 | — | — | — | — | — |
| 344 | — | — | 0.016 | — | — | — | — |
| 346 | — | 0.029 | — | — | — | — | — |
| 348 | — | — | — | — | 0.020 | 0.019 | 0.028 |
| 350 | — | — | — | — | 0.060 | — | — |
| N | 60 | 68 | 64 | 42 | 50 | 52 | 36 |
| TmoM11 | |||||||
| 147 | — | — | 0.155 | — | — | — | — |
| 149 | — | — | — | — | — | — | — |
| 151 | — | — | — | — | — | — | — |
| 153 | 0.200 | 0.118 | 0.034 | — | 0.020 | — | — |
| 155 | 0.083 | 0.118 | — | — | 0.040 | — | — |
| 157 | — | — | 0.431 | — | — | — | — |
| 159 | 0.633 | 0.676 | — | 0.476 | 0.060 | — | — |
| 161 | 0.017 | — | 0.259 | 0.452 | — | — | — |
| 163 | 0.017 | — | 0.017 | 0.071 | — | — | — |
| 165 | — | 0.029 | 0.017 | — | 0.560 | — | 0.056 |
| 167 | — | — | — | — | — | 0.981 | 0.917 |
| 169 | — | — | 0.086 | — | 0.100 | 0.019 | 0.028 |
| 171 | — | 0.015 | — | — | — | — | — |
| 173 | 0.050 | 0.029 | — | — | 0.220 | — | — |
| 175 | — | 0.015 | — | — | — | — | — |
| N | 60 | 68 | 58 | 42 | 50 | 52 | 36 |
| TmoM25 | |||||||
| 330 | 0.017 | — | — | — | — | — | — |
| 344 | — | — | — | 0.048 | — | — | — |
| 348 | — | 0.015 | — | — | 0.060 | — | — |
| 350 | 0.017 | 0.015 | — | 0.143 | — | 0.500 | 0.611 |
| 352 | 0.217 | 0.132 | — | 0.167 | — | 0.019 | — |
| 354 | 0.033 | 0.235 | — | 0.262 | 0.140 | 0.231 | 0.083 |
| 356 | 0.067 | 0.059 | 0.063 | 0.167 | 0.540 | — | — |
| 358 | 0.200 | 0.118 | 0.047 | 0.095 | — | 0.077 | — |
| 360 | 0.117 | 0.074 | 0.031 | 0.024 | 0.140 | 0.077 | — |
| 362 | 0.067 | 0.132 | 0.063 | 0.048 | 0.040 | — | 0.028 |
| 364 | 0.083 | 0.044 | 0.156 | — | 0.040 | 0.019 | 0.028 |
| 366 | 0.033 | 0.029 | 0.094 | — | — | — | — |
| 368 | — | 0.015 | 0.094 | 0.048 | — | 0.038 | 0.139 |
| 370 | 0.100 | 0.044 | 0.094 | — | — | — | — |
| 372 | 0.017 | 0.029 | 0.094 | — | — | — | — |
| 374 | — | 0.015 | 0.078 | — | — | 0.038 | 0.111 |
| 376 | — | — | 0.047 | — | — | — | — |
| 378 | 0.017 | 0.015 | 0.063 | — | 0.040 | — | — |
| 380 | — | 0.015 | 0.063 | — | — | — | — |
| 382 | — | — | 0.016 | — | — | — | — |
| 386 | — | 0.015 | — | — | — | — | — |
| 392 | 0.017 | — | — | — | — | — | — |
| N | 60 | 68 | 64 | 42 | 50 | 52 | 36 |
| Pzeb1 | |||||||
| 124 | 0.317 | 0.353 | 0.750 | — | 0.200 | 0.462 | 0.639 |
| 126 | 0.033 | 0.015 | 0.250 | — | 0.140 | 0.404 | 0.056 |
| 128 | 0.067 | 0.044 | — | — | — | — | — |
| 132 | 0.033 | — | — | 0.024 | — | — | — |
| 134 | 0.083 | 0.162 | — | 0.095 | — | — | — |
| 136 | 0.017 | 0.059 | — | 0.048 | 0.180 | — | — |
| 138 | 0.033 | 0.088 | — | — | 0.340 | 0.135 | 0.278 |
| 140 | 0.267 | 0.191 | — | 0.429 | — | — | 0.028 |
| 142 | — | 0.029 | — | 0.048 | — | — | — |
| 144 | 0.100 | 0.044 | — | 0.119 | 0.140 | — | — |
| 146 | 0.017 | — | — | 0.119 | — | — | — |
| 148 | — | — | — | 0.048 | — | — | — |
| 156 | — | — | — | 0.071 | — | — | — |
| 160 | 0.033 | — | — | — | — | — | — |
| 164 | — | 0.015 | — | — | — | — | — |
| N | 60 | 68 | 64 | 42 | 50 | 52 | 36 |
| Pzeb3 | |||||||
| 313 | 0.133 | 0.088 | 0.563 | — | 1.000 | 1.000 | 1.000 |
| 315 | 0.183 | 0.206 | 0.031 | 0.190 | — | — | — |
| 317 | 0.183 | 0.132 | 0.359 | 0.071 | — | — | — |
| 319 | 0.150 | 0.456 | — | — | — | — | — |
| 321 | 0.283 | 0.029 | 0.047 | 0.214 | — | — | — |
| 325 | 0.067 | 0.044 | — | 0.524 | — | — | — |
| 327 | — | 0.029 | — | — | — | — | — |
| 329 | — | 0.015 | — | — | — | — | — |
| N | 60 | 68 | 64 | 42 | 50 | 52 | 36 |
| UNH002 | |||||||
| 154 | 0.033 | — | — | — | — | — | — |
| 158 | 0.033 | 0.074 | — | — | — | — | — |
| 160 | — | 0.044 | 0.063 | — | 1.000 | 0.923 | 0.806 |
| 162 | 0.083 | 0.074 | — | 0.548 | — | 0.077 | 0.194 |
| 164 | 0.067 | 0.103 | — | — | — | — | — |
| 166 | 0.033 | 0.029 | 0.063 | 0.024 | — | — | — |
| 168 | 0.133 | 0.309 | 0.016 | 0.143 | — | — | — |
| 170 | 0.117 | 0.118 | 0.203 | 0.048 | — | — | — |
| 172 | 0.117 | 0.176 | 0.047 | 0.024 | — | — | — |
| 174 | 0.017 | — | 0.016 | — | — | — | — |
| 176 | 0.033 | — | 0.109 | — | — | — | — |
| 178 | 0.200 | 0.029 | 0.047 | 0.119 | — | — | — |
| 180 | 0.050 | 0.015 | 0.063 | 0.048 | — | — | — |
| 182 | 0.017 | — | 0.109 | — | — | — | — |
| 184 | — | — | 0.078 | — | — | — | — |
| 186 | 0.033 | 0.015 | 0.094 | — | — | — | — |
| 188 | — | 0.015 | 0.047 | 0.048 | — | — | — |
| 190 | — | — | 0.016 | — | — | — | — |
| 194 | — | — | 0.016 | — | — | — | — |
| 198 | 0.017 | — | — | — | — | — | — |
| 200 | 0.017 | — | — | — | — | — | — |
| 214 | — | — | 0.016 | — | — | — | — |
| N | 60 | 68 | 64 | 42 | 50 | 52 | 36 |
References
This study was part of Lukas Rüber’s PhD research applying molecular genetic techniques and morphometric analyses to study the evolutionary history of eretmodine cichlids from Lake Tanganyika. Axel Meyer is Professor in the Department of Biology at the University of Konstanz and interested in molecular phylogenetics and evolutionary biology. Christian Sturmbauer is Associate Professor at the Department of Zoology and Limnology at the University of Innsbruck and interested in molecular systematics and evolutionary processes in cichlids and a variety of other organisms. Erik Verheyen is a staff scientist at the Royal Belgian Institute of Natural Sciences in Brussels and is working on several projects involving the role of palaeoclimatic changes on speciation and evolution of selected African vertebrates. L.R is postdoctoral fellow in the laboratory of Rafael Zardoya at the Museo Nacional de Ciencias Naturales in Madrid.




