Expression of cell-associated prion protein on normal human platelets
The presence of normal prion protein (PrPC) within platelets was originally described by Perini et al (1996a) . Recently, Barclay et al (1999) reported the distribution of PrPC within normal human adult blood by flow cytometry. They calculated the amounts of PrPC on both leucocytes and platelets by comparison to bead standards. All mononuclear leucocyte subpopulations and platelets expressed PrPC, whereas both erythrocytes and polymorphonuclear leucocytes were virtually negative. Mononuclear cells expressed 3000–4000 molecules per cell. Activated platelets expressed approximately 4800 molecules per cell compared with 1400 molecules per cell on resting cells. The data agree fairly closely with the study by Holada et al (1998) with either ∼2000 or 4500 molecules of PrPC on resting and TRAP-activated platelets respectively. These results coupled with dissociation-enhanced fluoroimmunoassay (DELFIA) data from MacGregor et al (1999) demonstrate that platelets are the predominant source of peripheral blood prion protein.
We have also recently performed a similar flow cytometric study utilizing a commercial kit (Platelet PrP, Biocytex, Marseille, France). PrPC expression was measured on resting and activated platelets by flow cytometry and the number of molecules per platelet was calculated by utilizing the calibration beads within the kit. Platelets were identified according to their characteristic scatter profile and gating confirmed with anti-GpIb labelling, both pre- (97·5% ± 0·7%) and post-activation (96·5 ± 2·6%) with TRAP in separate tubes. The TRAP degranulation step was also confirmed with anti-CD62p in separate tubes, with resting and activated platelets expressing 3·6% (± 2·4%) and 93·6% (± 1·9) CD62p respectively. The prion protein results are summarized in Fig 1. Kit instructions, which do not involve any washing step, were followed almost exactly except that vortexing was avoided, as recommended in flow cytometric protocols for studying platelets ( Schmitz et al, 1998 ). The data suggested that resting platelets within PRP (n = 6) expressed approximately 297 ± 136 molecules of PrPC. Upon TRAP degranulation, the number of PrPC molecules increased to approximately 863 ± 223/platelet. The data take into account the subtraction of background labelling of both the isotype control antibody and secondary antibody, and agree closely with the expected figures (500–1500 molecules/platelet) quoted by the manufacturer. The PrPC signal was also sensitive to Proteinase K digestion, agreeing with the results of Barclay et al (1999) and Holada et al (1998) (results not shown). Although there is a quantitative discrepancy between our results and both those of Holada et al (1998) and Barclay et al (1999) , the results still suggest that platelets are the predominant pool of PrPC within whole blood. The differences between our results and Barclay et al (1999) or Holada et al (1998) could possibly be explained by a number of methodological issues. Firstly, the Biocytex test procedure is performed with a minimum saturating concentration of primary antibody and excess secondary antibody, and includes an isotype and concentration-matched negative control Mab to evaluate background fluorescence. Barclay et al (1999) used very high concentrations of primary antibody and no isotype control, but washed the platelets after the primary antibody. Although this may not be a large problem with platelets, this could potentially result in high background labelling of leucocytes. The possible non-specific binding of primary antibody, however, was controlled for by Holada et al (1998) without influencing the final copy number on platelets. Perhaps the discrepancy in the results is caused by the differences in utilization of calibration beads by the respective authors. As washing steps are avoided in the kit procedure, calibration beads and all kit reagents may have been optimized for use without washing. Interestingly, both Barclay et al (1999) and Holada et al (1998) used the same type of calibration bead preparations extracted from Biocytex Platelet kits in procedures involving washing steps. Indeed from the data presented in Fig. 4 of the paper from Barclay et al (1999) , very different fluorescence intensity levels can be observed on platelets [mean fluorescence intensity (MFI) < 10 arbitrary units (a.u.)] compared with lymphocytes (MFI > 60 a.u.). This could be consistent with a PrP copy number about five to ten times less on platelets than on lymphocytes and with our own data. Nevertheless, our study agrees with both Barclay et al (1999) and Holada et al (1998) in that platelets contain appreciable internal pools of PrPC. Preliminary transmission electron microscopy data suggest that intracellular PrPC is localized within platelet alpha granule membranes (data not shown) agreeing with the flow cytometric data of Holada et al (1998) . As platelets have been shown to contain PrPC mRNA ( Perini et al, 1996b ), it is highly probable that the platelet PrPC is derived from megakaryocyte synthesis.
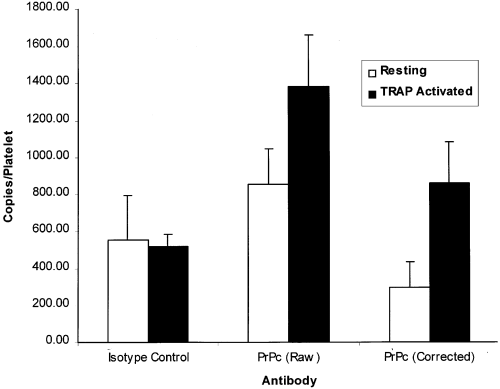
Surface expression of prion protein (PrPC) on normal human blood platelets (n = 6). Flow cytometry was performed on both resting and TRAP-activated platelets. The final corrected number of surface molecules per platelet was calculated by subtracting the background labelling of the isotype control antibodies from the raw data.




