Chromosome evolution in ground beetles: localization of the rDNA loci in the tribe Harpalini (Coleoptera, Carabidae)
Chromosomenevolution bei den Laufkäfern: Lokalisation der rDNA-Loci im Tribus Harpalini (Coleoptera, Carabidae)
Abstract
enThe pattern of localization of the ribosomal genes was studied by means of fluorescence in situ hybridization in 39 species of the tribe Harpalini. Most of them show one pair of autosomes carrying the ribosomal genes in a distal position of a single chromosome arm. This pattern is hypothesized to be ancestral for the whole tribe. Both, chromosome number and the number and localization of rDNA loci, show little variation and are therefore of little phylogenetic value. Only in the subtribe Ditomina is there enough variation to characterize phyletic relationships. The stability of rDNA loci is even higher than the constancy of chromosome number, as most species of Ditomina (genera Dixus, Eocarterus, Carterus, Odontocarus and Ditomus) have the usual pair of autosomes with rDNA loci, in spite of remarkable differences in the diploid number. Only Dixus sphaerocephalus and Dixus clypeatus have two autosomal pairs with a fluorescent signal. These results do not support the hypothesis that the high chromosome numbers found within Ditomina are the result of polyploid change from the ancestral 2n = 37 karyotype of the tribe Harpalini. Chromosomal translocations or the presence of mobile genetic elements are plausible sources of the few cases of intraspecific polymorphism in the rDNA loci found in species of Harpalus.
Zusammenfassung
deDas Muster der Lokalisation der ribosomalen Gene wurde bei 39 Arten des Tribus Harpalini mit Hilfe der Fluoreszenz-in situ- Hybridisierung untersucht. Bei den meisten von ihnen findet man ein Autosomenpaar, das die ribosomalen Gene in distaler Position an einem einzigen Chrosomenarm trägt. Dieses Muster wird als das hypothetisch ursprüngliche des ganzen Tribus angesehen. Beides, die Chromosomenzahl und die Zahl an lokalisierbaren rDNA-Loci, zeigen geringe Variation und sind deshalb von geringem phylogenetischen Aussagewert. Nur im Subtribus Ditomina gibt es genügend Abweichungen, aus denen sich phylogenetische Beziehungen ableiten lassen. Die Stabilität der rDNA-Loci ist sogar höher als die Konstanz der Chromosomenzahlen, insofern als die meisten Arten des Subtribus Ditomina (mit den Gattungen Dixus, Eocarterus, Carterus, Odontocarus und Ditomus), trotz beachtlicher Unterschiede in der diploiden Chrosomenzahl, das übliche Chromosomenpaar mit den rDNA-Loci haben. Nur Dixus sphaerocephalus und D. clypeatus besitzen zwei Chromosomenpaare mit einem fluoreszierenden Signal. Diese Resultate unterstützen die Annahme, daß die hohen Chromosomenzahlen, die beim Subtribus Ditomina gefunden wurden, nicht das Ergebnis einer polyploiden Veränderung des ursprünglichen Karyotyps 2n = 37 des Tribus Harpalini sind. Chromosomale Translokationen oder die Gegenwart von mobilen Elementen sind plausible Ursachen der wenigen Fälle eines intraspezifischen Polymorphismus für die rDNA-Loci, die bei Arten der Gattung Harpalus gefunden wurden.
Introduction
The tribe Harpalini Bonelli (Coleoptera: Carabidae) belongs to the phyletic lineage of carabid beetles with derived morphological characters (the Limbata Conchifera of Jeannel 1941, or the Harpalidae of Deuve 1988). To date, the systematics of the tribe has been based on morphological characters of adults and larvae (e.g. Csiki 1932; Emden van 1953; Noonan 1976), and also on male genitalia (Jeannel 1942). The tribe has been the object of different classifications according to the systematic importance given by different authors to particular morphological characters. Likewise, there are divergent criteria with respect to the division of Harpalini into subtribes and the limits of Harpalus Latreille and other complex genera.
The Ditomi ‘group’ was included by Noonan (1976) in the subtribe Harpalina, but its systematic position has changed following authors such as Jeannel (1942), Antoine (1959) and Lorenz (1998). The group comprises 16 genera and approximately 80 species with a turano-europeo-mediterranean pattern of geographical distribution; the Australian genus Phorticosomus Schaum should be excluded from Ditomina based both on molecular and morphological data (Martínez-Navarro, E.M.; Serrano, J.; Galián, J., unpublished results). The group should be better ranked as a separate subtribe, as it is well characterized by enough morphological and karyotypic autapomorphies (Martínez-Navarro et al. 2003), including a true presocial behaviour (Brandmayr and Zetto-Brandmayr 1987). Data on the sequence of the cytochrome-oxydase I corroborate the monophyly of the group (Fig. 2; Martínez-Navarro, E.M.; Serrano, J.; Galián, J., unpublished results).
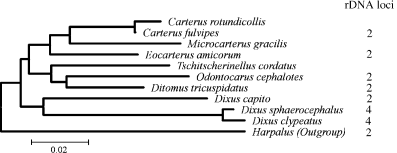
Phylogram based on the distance analysis (Kimura 2 parameters and neighbour joining) of a citochrome oxidase I fragment and number of rDNA loci detected (right) within the subtribe Ditomina group (Martínez-Navarro et al. unpublished results)
According to Noonan (1976), the genus Harpalus is the taxon with the highest number of species of the tribe, as it includes approximately 480 species belonging to 37 subgenera and six species groups. Harpalus is mainly concentrated in the Nearctic and Palearctic regions; there are 56 species in the Ethiopic region and a few in the Oriental region. Jeannel (1942), Antoine (1959), Vigna Taglianti (1993), Zaballos and Jeanne (1994) or Turin (2000), among other authors, proposed the fragmentation of Harpalus in more homogenous taxa, such as Harpalus s. str., Ophonus Dejean s. str., Hesperophonus Antoine, Cryptophonus Brandmayr & Zetto Brandmayr, Semiophonus Schauberger, Pseudoophonus Motschulsky, Platus Motschulsky or Pangus Motschulsky. Except for an initial cladistic analysis based on morphological characters of adults and larvae and karyotypic data (Martínez-Navarro et al. 2003), there are no explicit hypotheses about the relationships among subtribes of Harpalini or among genera within the subtribes.
The diploid chromosome number varies widely within the tribe, between 2n = 24 and 2n = 69 (data referred to 94 species; Serrano and Galián 1998). The karyotype with 2n = 37 chromosomes is the most frequent (more than 60 species), and is usually made up of metacentric and submetacentric chromosomes of medium-small size (1–4 μm). The karyotype with 2n = 37 chromosomes is considered the ancestral state for the tribe (Serrano et al. 1994), and also for the other tribes of the Caraboidea Conchifera stock (Serrano and Galián 1998). Most species of Harpalus and the closely related genera Ophonus s. str., Hesperophonus, Pseudoophonus and Cryptophonus have the same 2n = 37 diploid number (Serrano and Galián 1998). However, the subtribe Ditomina is characterized by high chromosome numbers between 2n = 41–69. It has been postulated that these high numbers are the result of a polyploidization followed by extensive karyotypic reorganization (Serrano et al. 1994). According to this hypothesis most loci should be detected in double amount within Ditomina. As the ribosomal genes (rDNA) in animals , 18S, 5,8S and 28S, form clusters of hundreds or thousands of repeated units (Hillis and Dixon 1991) spread along one or more chromosome pairs, they form good genetic marker to test such hypothesis. The chromosomal localization of these genes has already been the object of phylogenetic and evolutionary studies in beetle taxa of the two major suborders: Polyphaga (Juan et al. 1993) and Adephaga (Galián et al. 1995, 1999, 2002; Galián and Hudson 1999; De la Rúa et al. 1996; Sánchez-Gea et al. 2000; Proença and Galián 2003).
The study of the localization pattern of rDNA genes within Ditomina and other subtribes and genera of Harpalini may help in reconstructing the evolution of these genes within the tribe, and provide useful data for testing phylogenetic hypotheses derived from other character sets.
Materials and Methods
Material
Taxa and sampling localities of the specimens studied are listed in Tables 1–3. The material is deposited in the collection of the Department of Zoology and Physical Anthropology (University of Murcia, Spain).
| Species, num. individuals, sex | Locality | Diploid number and meioformula | Num. of rDNA loci |
|---|---|---|---|
| Harpalus distinguendus (Duft. 1812), 2 m | San Pedro del Pinatar (Murcia) | 37 (18 + X) | 4 |
| H. distinguendus (Duft. 1812), 1 m | Salinas de Pinilla (Albacete) | 37 (18 + X) | 4 |
| H. distinguendus (Duft. 1812), 1 m | Laguna del Salobralejo (Albacete) | 37 (18 + X) | 2 |
| H. contemptus Dej. 1829, 1 m | Cidones (Soria) | 37 (18 + X) | 2 |
| H. contemptus Dej. 1829, 1 m | Somosierra (Madrid) | 37 (18 + X) | 2 |
| H. microthorax salinator Mots. 1849, 1 m | Laguna de Peña Hueca (Toledo) | 37 (18 + X) | 2 |
| H. microthorax salinator Mots. 1849, 1 m | Laguna del Salobralejo (Albacete) | 37 (18 + X) | 2 |
| H. microthorax salinator Mots. 1849, 2 m | Salinas de Pinilla (Albacete) | 37 (18 + X) | 4 |
| H. affinis (Schr. 1781), 1 m | San Juan de los Abadeses (Gerona) | (19) | 2 |
| H. affinis (Schr. 1781), 1 m | Sierra de la Estrella (Portugal) | (19) | 2 |
| Harpalus sp., 1 m | SE Digor, Kars (Turkey) | ? | 2 |
| H. rubripes (Duft. 1812), 1 m | NW Dagpinar, Kars (Turkey) | 37 (18 + X) | 2 |
| H. rubripes (Duft. 1812), 1 m | El Hosquillo (Cuenca) | 37 (18 + X) | 2 |
| H. rubripes (Duft. 1812), 1 m | Popovi Livadi, South Pirin Mountains (Bulgaria) | 37 (18 + X) | 2 |
| H. serripes (Quens. 1806), 1 m | Las Majadas (Cuenca) | 37 (18 + X) | 2 |
| H. serripes (Quens. 1806), 1 m | Pto. de Menga (Ávila) | 37 (18 + X) | 2 |
| H. serripes (Quens. 1806), 1 m | Kozhukh Village, Petrich (Bulgaria) | 37 (18 + X) | 2 |
| H. anxius (Duft. 1812), 1 m | Cidones (Soria) | 37 (18 + X) | 2 |
| H. nevadensis Dan. 1898, 1 m | Sierra Nevada (Granada) | ? | 2 |
| H. honestus (Duft. 1812), 1 m | Javalambre (Teruel) | 37 (18 + X) | 2 |
| H. decipiens Dej. 1829 1 m | Pto. de Menga (Ávila) | 37 (18 + X) | 2 |
| H. decipiens Dej. 1829, 1 m | Sierra de Béjar (Salamanca) | 37 (18 + X) | 2 |
| *H. fuscipalpis Sturm 1825, 1 m | Villaverde (Albacete) | (19) | 4 |
| H. wagneri Schaub. 1926, 1 m | Valsaín (Segovia) | 30 (14 + XY) | 2 |
| H. wagneri Schaub. 1926, 1 m | Cidones (Soria) | 30 (14 + XY) | 2 |
| *H. rufipalpis Sturm 1818 1 m | Christo Smiznenski (Bulgaria) | 30 | 2 |
| Pseudoophonus rufipes (DeGeer 1774), 1 m | Tendire Pass, Agri (Turkey) | 37 (18 + X) | 2 |
| P. rufipes (DeGeer 1774), 1 m | Lestheri Village (Bulgaria) | 37 (18 + X) | 2 |
| *P. griseus (Panz. 1797), 1 m | Lestheri Village (Bulgaria) | 37 | 2 |
| P. (Platus) calceatus (Duft. 1812), 1 m | Jumilla (Murcia) | ? | 2 |
| Cryptophonus tenebrosus (Dej. 1829), 1 m | Fuente Álamo (Albacete) | 37 | 2 |
| Ophonus (Hesperophonus) azureus (F. 1755), 1 m | Erzincan (Turkey) | ca. 37 | 2 |
| *O. (Hesperophonus) cribricollis (Dej. 1829), 2 m | Gorno Spanchevo (Bulgaria) | ca. 37 | 2 |
| *O. (Hesperophonus) pumilio (Dej. 1829), 2 m | Pto del Viento (Málaga) | 37 (18 + X) | 2 |
| O.(Hesperophonus) pumilio (Dej. 1829), 2 m | Alozaina (Málaga) | 37 (18 + X) | 2 |
| O. (Ophonus) sabulicola hispanicus Schaub. 1926, 1 m | Erzincan (Turkey) | 37 | 6 |
| O. (Ophonus) ardosiacus Luts. 1922, 1 m | Mataporquera (Cantabria) | 37 (18 + X) | 2 |
| O. (Ophonus) ardosiacus Luts. 1922, 3 m | Villaverde (Albacete) | 37 (18 + X) | 2 |
| Species, num. individuals, sex | Locality | Diploid number and meioformula | Num. of rDNA loci |
|---|---|---|---|
| Harpalina | |||
| *Parophonus hispanus (Ramb. 1833), 2 m | Alozaina (Málaga) | ca. 37 | 2 |
| *P. hespericus Jeanne 1985, 1 m | Khénichét (Morocco) | (19) | 2 |
| *Acinopus picipes (Oliv. 1795), 1 m | Las Majadas (Cuenca) | 37 (18 + X) | 2 |
| Nesarpalus fortunatus (Woll. 1863), 1 m | Pozo de las Nieves (Gran Canaria) | 37 | 2 |
| Stenolophina | |||
| Egadroma piceus (Guérin-Mén. 1830), 1 m | Black Mountain, Canberra (Australia) | 37 (18 + X) | 4 |
| E. marginatum (Dej. 1829), 1 m | Salinas de Cordovilla (Albacete) | 39 (19 + X) | 2 |
| *Stenolophus abdominalis Géné 1836, 1 m | Moratalla (Murcia) | 37 (18 + X) | 2 |
| Dicheirotrichus obsoletus (Dej. 1829), 1 m | Cabo de Gata (Almería) | 37 | 2 |
| Pelmatellina | |||
| *Lecanomerus sp., 1 m | Black Mountain, Canberra (Australia) | 37 | 2 |
| Species, num. individuals, sex | Locality | Diploid number and meioformula | Num. of rDNA loci |
|---|---|---|---|
| Dixus sphaerocephalus (Oliv. 1795), 1 m | Pto.Viento, Ronda (Málaga) | 55 (27 + X) | 4 |
| D. sphaerocephalus (Oliv. 1795), 1 m | San Pedro del Pinatar (Murcia) | 55 (27 + X)’ | 4 |
| D. sphaerocephalus (Oliv. 1795), 2 m | Villaverde (Albacete) | 55 (27 + X) | 4 |
| D. clypeatus (Rossi 1790), 1 m | Casarabonela (Málaga) | 45 (22 + X) | 4 |
| D. clypeatus (Rossi 1790), 1 m | Sierra del Relumbrar (Albacete) | 45 (22 + X) | 4 |
| D. capito (Aud.-Serv. 1821), 1 m | Pto Viento, Ronda (Málaga) | 69 (34 + X) | 2 |
| Odontocarus cephalotes (Dej. 1826), 1 m | Pto Viento, Ronda (Málaga) | ca. 40 | 2 |
| Carterus fulvipes (Latr. 1817), 1 m | El Bonillo (Albacete) | 57 | 2 |
| *Eocarterus amicorum Wrase 1993, 1 m | Ronda (Málaga) | ca. 40 + X | 2 |
| E. amicorum Wrase 1993, 1 m | Pto Viento, Ronda (Málaga) | ca. 40 + X | 2 |
| Ditomus tricuspidatus (F. 1792), 1 m | Pto. Viento, Ronda (Málaga) | 59 | 2 |
Chromosome preparations
Karyotypic observations were made on cells from male adult gonads. The testes were removed from beetles anaesthetized with ethyl acetate, treated hypotonically with distilled water, fixed in fresh ethanol–acetic acid solution (3 : 1), and stored at −20°C until study. Small sections of the gonads were squashed on a slide in 70% acetic acid and the coverslip was removed after freezing in liquid nitrogen. The slides were dried on a 40°C hot plate and stored at −20°C until in situ hybridization.
DNA probe
Total DNA from specimens of the genus Harpalus was extracted using the Chelex protocol (Walsh et al. 1991). The ribosomal probe used was an 18S fragment of 555 bp amplified by polymerase chain reaction (PCR) with the fungus primers NS1 and NS2 (White et al. 1990). The probe was labelled in a second PCR with biotin-16-dUTP.
In situ hybridization
Chromosome preparations were pretreated with glacial acetic acid for 60 min, digested with RNAse I in 2 × SSC (100 μg/ml) for 1 h at 37°C and with pepsin in 0.01 M HCl (0.7 μg/ml) for 10 min at 37°C. After digestion the chromosomes were fixed with 4% fresh paraformaldehyde in NaOH 0.1 N, dehydrated in a graded ethanol series and air-dried. The hybridization mixture was prepared as described in Sánchez-Gea et al. (2000). The labelled probe was denatured at 100°C during 5 min and placed on ice. The slides were heated on a 80°C hot plate for 5 min. The denatured hybridization mixture was placed over the denatured slides and covered with a coverslip. The slides were then transferred to a humid chamber at 80°C, allowing temperature to drop slowly to 37°C for hybridization overnight. The slides were washed following the protocol described in Sánchez-Gea et al. (2000). Hybridization sites were detected with avidin-fluorescein isothiocyanate (FITC), and the signal was amplified twice using goat biotinylated antiavidin. The chromosomes were counterstained with propidium iodide and the slides were mounted with antifade solution. Photographs were made with a standard colour negative film of 100 ASA, or images were captured with a video camera connected to the photomicroscope.
Results
Localization and number of rDNA loci in Harpalini
The localization of the ribosomal genes has been determined in 39 species (Tables 1–3). Most species show two autosomes with ribosomal genes in mitotic metaphase (MM) (Fig. 1a), or one bivalent in metaphase I (MI) (Fig. 1b), two hybridization marks in interphase nuclei (Fig. 1c), or one mark in interkinesis (Fig. 1d) and spermatid nuclei. Hybridization sites are distally located in a single arm of mitotic chromosomes (Fig 1a). Variations of this pattern are found in species of the genus Harpalus as an intraspecific polymorphism, which perhaps has been fixed in Harpalus fuscipalpis Sturm (Table 1). One individual of Harpalus distinguendus Duftschmid collected in Salinas de Pinilla presents signals in two autosomal pairs (Fig. 1e), same for two individuals from San Pedro del Pinatar, whereas the population from Laguna del Salobralejo shows hybridization signals only in one autosomal pair (Fig. 1f). One individual of Harpalus microthorax Motschulsky from Peña Hueca and one from Laguna del Salobralejo show the normal pattern described above, whereas two individuals from Salinas de Pinilla show four hybridization sites. One species of Ophonus has six rDNA loci, and interspecific differences in the number of rDNA loci are found in Egadroma (Table 2; Fig. 1g) and Dixus (Table 3; Fig. 1h and i). In the case of Harpalus wagneri Schauberger and Harpalus rufipalpis Sturm, which have a XY sex chromosome system, the lack of a fluorescent mark in the sex vesicle suggests that rDNA clusters are located in one autosomal pair. In species such as Harpalus serripes Quensel the size of the fluorescent signal in mitotic chromosomes is variable among homologues of the same individual.
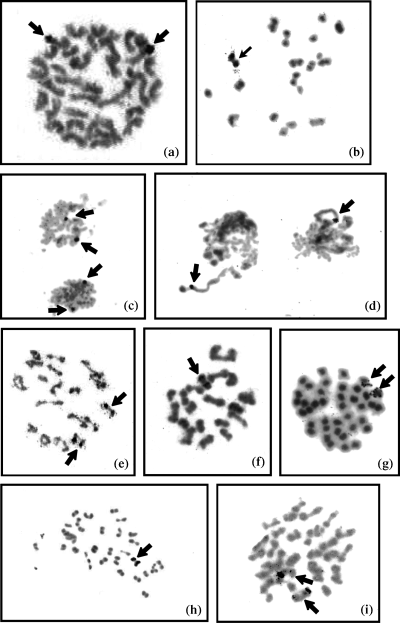
Localization of rDNA loci by FISH of a PCR-amplified 18S probe to spermatogonial and nuclei from species of the tribe Harpalini. (a) Ophonus pumilio (2n = 37, 18 + X), mitotic metaphase plate with two labelled chromosomes. (b) Harpalus rubripes, (2n = 37, 18 + X) metaphase I with one labelled bivalent. (c) Pseudoophonus griseus (2n = 37), interphase nuclei with two fluorescent signals each. (d) Pardileus calceatus (2n = ?), interkinesis nuclei with one fluorescent signal each. (e) Harpalus distinguendus (2n = 37, 18 + X) (Laguna del Salobralejo population), metaphase I showing one labelled bivalent. (f) H. distinguendus (2n = 37, 18 + X) (Salinas de Pinilla population), metaphase I showing two labelled bivalents. (g) Egadroma marginatum (2n = 39), mitotic metaphase showing two labelled chromosomes. (h) Dixus capito (2n = 69, 34 + X) showing one labelled bivalent. (i) Dixus sphaerocephalus (2n = 56, 22 + X) showing two labelled bivalents
Most species of the subtribe Ditomina (2n = 41 − 2n = 69) show the same pattern of two hybridization marks in interphase nuclei, one hybridization mark per spermatid or one labelled bivalent in MI (Table 3). This same pattern has been observed in Dixus capito Serville (Fig. 1h), but Dixus sphaerocephalus Olivier (2n = 55; n = 27 + X; Fig. 1i) and Dixus clypeatus Rossi (2n = 45) have two bivalents carrying the ribosomal genes. In one mitotic pair of this last species, the rDNA clusters occupy almost the whole arm, whereas in the second pair there is only a telomeric signal.
Chromosome number
The results corroborate previous data on species of Harpalini. The results on 11 species are new for the tribe (Tables 1–3). Most of them have a 2n = 37 karyotype, a result that corroborates the numerical stability shown by most lineages of Harpalini.
Discussion
The number and localization of rDNA sites seem to follow a generalized pattern in the tribe Harpalini, as there is usually only one autosomal pair with a fluorescent signal distally located in one chromosome arm. This pattern has been found in the tribe Carabini (De la Rúa et al. 1996) but is more variable in the tribes Scaritini (Galián et al. 1999), Zabrini (Sánchez-Gea et al. 2000) and Cicindelini (Galián et al. 1995, 2002; Proença and Galián 2003).
The stability of this pattern is perhaps because of the factors that maintain the karyotype with 2n = 37 chromosomes in most lineages of the tribe. However, some other factors must be involved, as the pattern is also found in the numerically variable Ditomina. The remarkable stability in both the diploid number and the number and localization of rDNA sites makes these characters useless for phylogenetic analyses at most taxonomic ranks within the Harpalini.
The occurrence of the generalized pattern for rDNA sites in the genera Dixus, Carterus, Eocarterus, Odontocarus and Ditomus, does not support the hypothesis of a polyploid change at the origin of the high chromosome numbers found in species of Ditomina. According to this hypothesis, the 2n = 37 karyotype of the ancestral Ditomine was duplicated into a 2n = 74 karyotype, a change followed by many others that lead to the remarkable variation in chromosome number of the group. Therefore, it was expected to find four rDNA loci in most species of this taxon.
The only variation within the Ditomina in the pattern of rDNA loci has been found in the genus Dixus. Whereas D. capito (2n = 69) has the usual single autosomal pair with ribosomal loci, D. sphaerocephalus (2n = 55) and D. clypeatus (2n = 45) have two pairs of autosomes showing a fluorescent signal. This result supports a close relationship between the last two species, which also seem to be closely related, based on morphological and molecular data (Fig. 2; Martínez et al. unpublished results).
The variations in the number of autosomes bearing rDNA loci (4 or 6) found in some species of the tribe, should be considered as a derived state caused by unknown chromosome rearrangements. Either reciprocal translocations or transposons may have originated the increase in the number of rDNA sites. The presence of transposons is well known in Drosophila (e.g. Daniels et al. 1990), Hymenoptera (Bigot et al. 1992) and other insect groups including Coleoptera (Burke et al. 1993). These transposable genetic elements have been considered to be a plausible explanation for the numerical variation of rDNA sites in the genus Zabrus (up to 12 chromosomes may show fluorescent signals; Sánchez-Gea et al. 2000), a taxon not distantly related to Harpalini. Interestingly, two of the cases of intraspecific polymorphism have been found in species of Harpalus collected in the same locality (Salinas de Pinilla, Albacete). This finding suggests a case of horizontal transmission of transposable elements between related species. These carabids frequently have parasitic mites (personal observations), that have been considered as potential vectors of transposable elements between species of Drosophila (Houck et al. 1991).
In most species, the ribosomal clusters occupy a distal position in one chromosome arm. This localization of rDNA genes has already been reported in carabids such as Cychrus caraboides Linne (De la Rúa et al. 1996) and the Cicindelini (Galián and Hudson 1999). In some species the size of the fluorescent region varies between homologues. This suggests the existence of a polymorphism for the number of copies of the ribosomal genes, as described by Sánchez-Gea et al. (2000) in the carabid genus Zabrus. These cases of polymorphism do not contradict the high uniformity shown by rDNA loci in Harpalini, which is probably the result of a concerted evolution discussed in detail by Hillis and Dixon (1991).
Acknowledgements
This work was supported by the projects PB98-0402 and BOS2002-02870 of the Spanish Ministry of Science and Technology. E. M. Martínez-Navarro was supported by a PhD studentship of the University of Murcia. Thanks are due to Dr. B. Moore, Dr. A. Slipinsky, Dr. R. Oberprieler, Dr. D. Yeates, Dr. D. Rentz,, Dr. L. Mound and T. Weir for their help in collecting Australian beetles. Prof. A. Kocak and Dr M. Kemal helped in collecting Turkish species, Dr. B. Guéorguiev with Bulgarian species, and Dr. A. Andújar, J. L. Lencina and J. F. Sánchez-Gea with Spanish species. Dr. B. Fernández is also acknowledged for providing laboratory facilities.




