Susceptibility gene for non-obstructive azoospermia in the HLA class II region: correlations with Y chromosome microdeletion and spermatogenesis
Summary
We previously reported an association between the human leukocyte antigen (HLA) haplotype DRB1*1302-DQB1*0604 in the HLA class II region and non-obstructive azoospermia in Japanese men. To identify possible associations between the HLA-DRB1*1302-DQB1*0604 allele in the HLA class II region and azoospermia factor (AZF) deletion in the Y chromosome, we performed genomic polymerase chain reaction (PCR) analysis of the AZF region. We then determined spermatogenic impairment (Johnsen score) in testicular biopsy specimens from patients with or without the DRB1*1302-DQB1*0604 haplotype. The AZF microdeletion rate in patients with this haplotype was 3.85%, compared with 11.8% in others (no correlation). However, Johnsen scores in patients with the DRB1*1302-DQB1*0604 haplotype were 3.13 ± 1.34 (mean ± SD), compared with 3.70 ± 1.51 in others (p < 0.05). While the DRB1*1302-DQB1*0604 haplotype acts independently from Y chromosome deletion, the haplotype might either act directly, or be functionally related to an unknown autosomal gene. In either case, this haplotype showed association with severe spermatogenic impairment.
Introduction
Infertility and subfertility have become a major clinical and social problem, affecting 13% of couples attempting to conceive their first child (Greenberg et al., 1978). The proportion of men with idiopathic infertility may be as high as 66%, depending on the definition of ‘idiopathic’ (Dubin & Amelar, 1971; Hendry et al., 1973; Greenberg et al., 1978). The aetiology of the disorder is not well understood. In particular, little is known about possible contributions of genetic factors (Mak & Jarvi, 1996; Reijo et al., 1996).
Y chromosome microdeletions at the azoospermia factor (AZF) locus have been implicated as a major genetic cause among cases of idiopathic male infertility (Liow et al., 2001). About 13% of occurrences of non-obstructive azoospermia result from deletions involving the AZF (Reijo et al., 1996). Vogt et al. (1996) reported three different AZF regions, AZFa, AZFb and AZFc. Candidate genes have been isolated for most AZF loci, including USP9Y (Homo sapiens chromosome Y ubiqutin-specific protease 9) (Brown et al., 1998) for AZFa; the RNA binding motif (RBMY) gene family for AZFb (Ma et al., 1993); and the deleted in azoospermia (DAZ) gene cluster for AZFc (Reijo et al., 1995). AZFd region also has been proposed by Kent-First et al. (1999). AZFd has been localized between AZFb and AZFc, but a candidate gene has not yet been found (Kent-First et al., 1999). In more than half of cases of non-obstructive azoospermia, deletion at a Y chromosome locus cannot be demonstrated. Loci other than AZF on the Y chromosome may be responsible.
We previously reported that human leukocyte antigens (HLA)-A33 and -B44 in the HLA class I region (Miura et al., 1998), and HLA-DRB1*1302 in the HLA class II region (Tsujimura et al., 1999), are linked to susceptibility to non-obstructive azoospermia in Japanese men. Moreover, certain microsatellite markers in the HLA class II region and at HLA-DRB1 and -DQB1 loci displayed strong associations with non-obstructive azoospermia (Tsujimura et al., 2002b). In particular, the frequency of the HLA-DRB1*1302-DQB1*0604 allele was increased in Japanese men with non-obstructive azoospermia. HLA classII (DR, DQ, DP) genes are characterized by extraordinary polymorphism and strong linkage disequilibrium between alleles at the DR and DQ loci. For example, DRB1*1302 is associated with DQB1*0604 in Japanese and also Norwegian populations (Yasunaga et al., 1996). We suggested two reasons that might account for the high frequency of the HLA-DRB1*1302-DQB1*0604 allele in Japanese men with non-obstructive azoospermia. First, a critical region for development of non-obstructive azoospermia might be located near the HLA-DRB1 and –DQB1 segments in the HLA class II region, and would show strong linkage to the HLA-DRB1 and –DQB1 segments. However, we have been unable to find a ‘candidate’ gene near the HLA region. Alternatively, HLA-DRB1 and –DQB1 segments might be linked to a gene on another chromosome. Among possible loci, only the AZF region is known to be a responsible for some cases of azoospermia. In our present examination of linkage of HLA-DRB1*1302-DQB1*0604 in the HLA class II region to AZF deletion in the Y chromosome, we first performed genomic polymerase chain reaction (PCR) to detect deletion in the AZF region of the Y chromosome. We then compared the deletion rate between patients with and without the HLA-DRB1*1302 -DQB1*0604 haplotype. Next we investigated the histopathology of spermatogenesis in testicular biopsy specimens to determine whether HLA-DRB1*1302 -DQB1*0604 in the HLA class II region was a risk factor for spermatogenic impairment.
Materials and methods
Subjects
We studied 60 infertile Japanese men with non-obstructive azoospermia (Tsujimura et al., 2002b). Diagnosis was confirmed by findings at vasography together with a Johnsen score (JSC) of <7 in a testicular biopsy specimen (Johnsen, 1970). We excluded from study any patient with chromosomal abnormalities or obstructive azoospermia. All samples were collected after obtaining informed consent. Based on previous HLA typing, we divided the 60 patients into two groups, 24 with HLA-DRB1*1302-DQB1*0604 in the HLA class II region and 36 patients with another haplotype.
Histopathology
Testicular specimens were evaluated based on the most advanced pattern of spermatogenesis present, e.g. hypospermatogenesis (HS; reduction in the degree of normal spermatogenesis, and focal abnormal spermatogenesis showing spermatid stage arrest); maturation arrest (MA; absence of any germ cells in the later stages of spermatogenesis); and Sertoli cell only syndrome (SCO; absence of any germ cells in seminiferous tubules) (Tsujimura et al., 2002a). In addition, the JSC was determined in each case.
Genomic DNA extraction
Genomic DNA was extracted using a salting-out procedure after treatment of peripheral blood leukocytes with proteinase K, as described elsewhere (Miura et al., 1998; Tsujimura et al., 1999; Tsujimura et al., 2002b).
PCR
To detect deletions from the Y chromosome, we performed PCR using the Y Chromosome Deletion Detection System, Version 1.1 (Promega, Madison, WI, USA) according to the manufacturer's instructions. In brief, this system consists of 18 primer pairs homologous to previously identified and mapped sequence-tagged sites (STS). The primers are combined into four sets for use in multiplex PCR, permitting assessment of all 18 STS by only four PCR amplifications. Each multiplex primer set includes a control primer pair that amplifies a fragment of the X-linked SMCX locus, serving as an internal control for the amplification reaction and integrity of the genomic DNA sample. Genomic DNA (250 ng) was used for PCR with a Takara thermal cycler. Conditions for all PCR reactions were as follows: denaturation at 94 °C, 1 min; annealing at 57 °C, 30 s; and extension at 72 °C, 1 min. After 35 cycles, a final extension was carried out at 72 °C for 5 min. After electrophoretic separation in a 4% agarose gel, we examined appropriate bands using ethidium bromide for visualization.
Statistical analysis
We evaluated the significance of differences by the chi-squared test and an umpaired t-test. When the expectation value was five or less, a p-value was determined using Fisher's direct probability method.
Results
AZF deletion
We detected Y chromosome deletions in five men in our genomic PCR analyses of the AZF region (Fig. 1). Six, six, five and five bands including the X-linked SMCX locus were detected with the respective primer mixtures A, B, C and D in a positive control (lanes 1, 5, 9 and 13). Lanes 2 and 3 exhibited three (SY254, SY157, SY130) and one (SY157) deletions, respectively; lanes 6 and 7, three (SY242, SY239, SY208) and five (SYPR3, SY127, SY242, SY239, SY208) deletions; lanes 10 and 11, one (SY255) deletion each; lanes 14 and 15, two (SY152, SY153) and three (SY152, SY153, SY124) deletions. Genetic findings in all patients with deletions from the AZF region are summarized in Fig. 2. The deletion rate for the total number of patients was about 8.33% (Table 1). The microdeletion rate for the AZF region in patients with the DRB1*1302/DQB1*0604 haplotype was about 3.85%, compared with 11.8% in other patients. Although there is a small number of patients, no statistically significant correlation was apparent between AZF deletion and haplotype DRB1*1302/DQB1*0604 (p = 0.377).
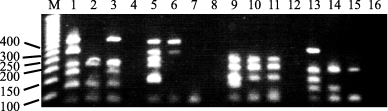
Y chromosome deletion analysis. Lanes 1–4 showed the amplification products from reactions using Multiplex A Master Mixture with a positive male genomic DNA control (lane 1), genomic DNA from a male with a Y chromosome deletion (lanes 2 and 3) and no DNA (lane 4). Lanes 5–8 show the amplification products from reactions using Multiplex B Master Mixture with a positive male genomic DNA control (lane 5), genomic DNA from a male with a Y chromosome deletion (lanes 6 and 7) and no DNA (lane 8). Lanes 9–12 show the amplification products from reactions using Multiplex C Master Mixture with positive male genomic DNA control (lane 9), genomic DNA from a male with a Y chromosome deletion (lanes 10 and 11) and no DNA (lane 12). Lanes 13–16 show the amplification products from reactions using Multiplex D Master Mixture with positive male genomic DNA control (lane 13), genomic DNA from a male with a Y chromosome deletion (lanes 14 and 15) and no DNA (lane 16). Markers (M) are the 50 bp DNA Step Ladder. The reactions were performed on Takara thermal cycler. The gel is a 4% agarose gel.

Deletion mapping of the AZF region on human Y chromosome in non-obstructive azoospermic men with and without the DRB1*1302/DQB1*0604 haplotype. Y chromosomal STS and locus names are listed on top border. Solid black box and (−) showed the presence and absence of STS.
| Haplotype DRB*1302/ DQB1*0604 | No. of patients | No. of microdeletions (%) |
|---|---|---|
| + | 26 | 1 (3.85) |
| − | 34 | 4 (11.8) |
| Total | 60 | 5 (8.33) |
Histopathology
The JSC for testicular biopsy specimens (Table 2) was lower for patients with the haplotype DRB1*1302/DQB1*0604 (3.13 ± 1.34; mean ± SD) than for other patients (3.70 ± 1.51) (p < 0.05). In patients with the DRB1*1302/DQB1*0604 haplotype, SCO was the predominant histologic diagnosis (73%, compared with HS in 11.5% and MA in 15.4%). SCO was more common in these patients than in those without DRB1*1302/DQB1*0604.
| Haplotype DRB1*1302 /DQB1*0604 | JSC (mean ± SD) | HS (%) | MA (%) | SCO (%) | Total number |
|---|---|---|---|---|---|
| + | 3.13 ± 1.34* | 3 (11.5) | 4 (15.4) | 19 (73.0) | 26 |
| − | 3.70 ± 1.51* | 10 (29.4) | 6 (17.6) | 18 (52.9) | 34 |
| 60 |
- JSC, Johnsen score; HS, Hypospermatogenesis; MA, Maturation arrest; SCO, Sertoli cell only. *p < 0.05.
Discussion
Male infertility remains idiopathic in most cases. Little is known about possible contributions of genetic factors (Mak & Jarvi, 1996; Reijo et al., 1996). Knockout mice lacking various functioning genes have exhibited disordered spermatogenesis and spermiogenesis (Blendy et al., 1996; Nantel et al., 1996; Zhang et al., 2001). Human homologues of such genes may prove to be responsible for some cases of oligozoospermia or azoospermia.
One major gene associated with male infertility has been termed the AZF. Microdeletion frequency at this locus has a reported range of 2.7–55.5% in patients with this problem (Kent-First et al., 1999). The microdeletions rate of AZFc region has been reported 6.7% in infertile Japanese men (Sawai et al., 2002). These deletion rates in azoospermia and severe oligozoospermia have varied widely reflecting differences in number of patients and number of STS markers studied. Simoni et al. (1999) have proposed the guidelines for standardization of screening for microdeletions of the Y chromosome (Simoni et al., 1999). We performed PCR using a kit including 18 STS. The total rate of microdeletions was 8.33% comparable with results in similar studies.
The Y chromosome deletions were first to be studied in azoospermic patients. Because deletion of both alleles on autosomal chromosomes is rare, expression of an abnormal phenotype can be difficult to detect even when a deleterious gene abnormality is present on one autosome. We examined HLA genes with respect to male infertility as an initial step in characterizing the roles of autosomes. In the mouse, the T/t alleles are located within the murine homologue of the major MHC on chromosome 17, and several genes expressed during spermatogenesis have been cloned within these regions (Lader et al., 1989; Ha et al., 1991; Mazarakis et al., 1991). Others have suggested an association of certain HLA molecules in spermatozoa (Martin-Villa et al., 1999), and also of the HLA class II region (van der Ven et al., 2000), with male infertility. The latter group found DRB1-DQA1-DQB1 haplotypes 0101-0101-0501 and 0701-0201-0201/0303 to be increased in patients with severe male-factor infertility, suggesting that they might confer susceptibility to this disorder. On the other hand, we previously reported that DRB1*1302/DQB1*0604 displayed strong associations with non-obstructive azoospermia in Japanese men. The difference in findings between that study and the findings of Van der Ven were thought to reflect differences in patient ethnicity and selection criteria. Our subjects all had non-obstructive azoospermia, while many of their subjects retained some capacity to produce sperm. The ethnic and geographic differences would be reflected in HLA haplotypes. We suggested that in a Japanese population a critical region for development of non-obstructive azoospermia linked to DRB1*1302/DQB1*0604 in the HLA class II region.
We investigated linkage of the above HLA haplotype to AZF deletion in the Y chromosome, as we were uncertain whether the HLA DRB1*1302/DQB1*0604 haplotype affected spermatogenesis independently or via an associated AZF deletion. The microdeletion rate in the AZF region in patients with this haplotype was about 3.85%, compared with 11.8% in other subjects (no correlation), suggesting that the haplotype acted independently of AZF. Further, as we found only a small number of subjects with deletions, we could not evaluate any tendency of the length of these deletions to reflect presence or absence of DRB1*1302/DQB1*0604. Further studies are needed to determine whether HLA influences azoospermia directly or via linkage to other, presently unknown, autosomal genes.
We analysed histologic features of biopsy specimens to determine whether HLA DRB1*1302/DQB1*0604 haplotype could serve as a marker for testicular germ cell development. The mean JSC of patients with the DRB1*1302/DQB1*0604 haplotype was lower than that of patients without this haplotype (p < 0.05), associating the haplotype with less favourable morphology. In patients with the DRB1*1302/DQB1*0604 haplotype, SCO was the most common finding (73%) to HS (11.5%) and MA (15.4%), representing a higher prevalence of SCO than that seen in other patients. Although various severities of defects in spermatogenesis were seen with the DRB1*1302/DQB1*0604 haplotype, specific defects in spermatogenesis cannot yet be linked with a specific gene. Although the AZFa region generally was associated with less favourable testicular histopathology, germ cells and even mature sperm occasionally were present (Kent-First et al., 1999). Recent demonstration of immunohistochemical phenotypes in association with molecular defects may be important for understanding normal and abnormal spermatogenic mechanisms (Blagosklonova et al., 2002). Nevertheless, cytokeratin-18 antibody (Bar-Shira Maymon et al., 2000) and two potential markers for Sertoli cell functional status (anti-Mullerian hormone or AMH, and M2A) (Blagosklonova et al., 2002) did not show any difference in immunoreactivity between AZF-deletion-patients and others. In the future we plan to examine immunoreactivity for these proteins in our patients with and without the DRB1*1302/DQB1*0604 haplotype.
In conclusion, we showed that the DRB1*1302-DQB1*0604 haplotype influences spermatogenesis independently of Y chromosome deletions, and might be linked to a novel autosomal azoospermic gene. The DRB1*1302-DQB1*0604 haplotype may be associated with severe spermatogenic impairment.
Acknowledgements
We thank Miss K. Adachi, M. Omune and M. Ohki for excellent technical assistance, and Dr M. Ota for an important discussion.




