Molecular characterization of the 3-phosphoglycerate kinase gene (PGK1) from the methylotrophic yeast Pichia pastoris
Abstract
We report the cloning of the 3-phosphoglycerate kinase gene (PGK1) from the methylotrophic yeast Pichia pastoris by a PCR approach. The coding sequence of the PGK1 gene comprises 1251 bp with the potential to encode a polypeptide of 416 amino acid residues, which shows high identity to homologous proteins from other yeasts. The promoter region of this gene (PPGK1) contains regulatory cis-elements found in other PGK1 genes, such as TATA box, CT-rich block and a heat shock element. In the 3′ downstream region we identified a tripartite element 5′-TAG–TAGT–TTT-3′, which is supposed to be important for transcription termination. As in other yeasts, the PGK1 gene from P. pastoris is present as a single-copy gene. Northern blot analysis revealed that the gene is transcribed as a 1.5 kb mRNA; when cells are grown on glucose the levels of this mRNA are increased two-fold in comparison to cells grown on glycerol. The transcriptional regulation of this gene by the carbon source was further confirmed when the α-amylase gene from Bacillus subtilis was placed under the control of PPGK1: higher levels of expression were obtained when cells were grown on glucose as compared to glycerol and methanol. Preliminary results related to the strength of PPGK1 show that it represents a potential alternative to constitutive heterologous expression in P. pastoris. The sequence of the gene has been deposited in GenBank under Accession No. AY288296. Copyright © 2005 John Wiley & Sons, Ltd.
Introduction
Genes from the glycolytic pathway are known to be highly expressed and the corresponding enzymes may represent 30–60% of the total soluble proteins in Saccharomyces cerevisiae (Fraenkel, 1982). 3-Phosphoglycerate kinase (PGK) is a glycolytic enzyme, which transfers a phosphoryl group from 1,3-bisphosphoglycerate to ADP, forming ATP and 3-phosphoglycerate. This enzyme also acts in the gluconeogenic pathway and may represent about 5% of the total cellular protein (Holland and Holland, 1978). So far, the gene that codes for this enzyme (PGK1) has been isolated from 47 species (Fleming and Littlechild, 1997). Due to the high levels of expression of the PGK1 gene, its promoter has been used in the construction of expression vectors for several yeasts and filamentous fungi: S. cerevisiae (Tuite et al., 1982; Mellor et al., 1983; Kingsman et al., 1985; Grange et al., 1996), Yarrowia lipolytica (Dall et al., 1996), Candida maltosa (Masuda et al., 1994), Kluyveromyces lactis (Fournier et al., 1989), Penicillium citrinum (Nara et al., 1993), Trichoderma reesei (Vanhanen et al., 1989, 1991), Rhizopus niveus (Takaya et al.,1994) and Aspergillus nidulans (Streatfield et al., 1993).
In S. cerevisiae, several regulatory elements have been identified in the 5′-promoter region of the PGK1 gene. An upstream activating sequence (UAS) localized between positions −538 to −402 (Stanway et al., 1987) has been implicated in the response mediated by the regulator Rap1p (Stanway et al., 1987; Chambers et al., 1989). Another important cis-element is formed by three repeats of the pentamer 5′-CTTCC-3′, which makes up the binding site for the glycolysis specific factor Gcr1p (Baker, 1991). The interaction between Gcr1p and its responsive element site is essential for complete promoter activation and contributes to specific activation mechanisms of glycolytic genes through the recruitment of Gcr2p (Zeng et al., 1997; Deminoff and Santangelo, 2001). Another cis-acting element present on the PGK1 promoter is the heat-shock element (HSE), which allows an increase of mRNA levels upon a shift to higher temperatures (Piper et al., 1988). In addition, a TATA box element has been identified, which is responsible for correct transcription initiation but does not influence the high expression levels of the PGK1 gene (Ogden et al., 1986; Rathjen and Mellor, 1990).
A downstream activating sequence (DAS) has been reported within the coding sequence of the PGK1 gene. As in S. cerevisiae, the PGK1 coding sequence from Y. lipolytica exerts a positive effect in heterologous expression under the control of its promoter, but this increase of expression is dependent on growth conditions (Dall et al., 1996). The absence of a DAS in expression vectors can explain in part the low yield of foreign proteins expressed under control of the PGK1 promoter when compared to the endogenous levels of 3-phosphoglycerate kinase (Mellor et al., 1987).
Expression studies with the PGK1 promoter have demonstrated a moderate regulation by the carbon source. In Aspergillus oryzae, a low variation of mRNA levels was observed when the carbon source was glucose or glycerol, but the mRNA levels decreased when the carbon source was pyruvate (Nakajima et al., 2000). The same pattern of expression was observed in S. cerevisiae; higher levels of mRNA when cells were grown in glycolytic substrates (glucose or glycerol) as opposed to gluconeogenic ones (pyruvate or acetate) (Stanway et al., 1987). In Y. lipolytica and A. nidulans, an opposite effect was observed; higher levels of PGK mRNA in gluconeogenic substrates as compared to glycolytic substrates (Dall et al., 1996; Streatfield et al., 1992). In Candida maltosa, the PGK1 gene was highly expressed by glucose-grown cells, whereas its mRNA could not be detected when cells were grown in n-tetradecane or n-tetradecanol (Masuda et al., 1994)
In the last 15 years, the methylotrophic yeast Pichia pastoris has been used with great success as an expression system in which approximately 300 different foreign proteins have already been expressed (Cereghino and Cregg, 2000). Most of the expression vectors for P. pastoris use the inducible promoter from the alcohol oxidase gene (AOX1), which codes for the first enzyme in the methanol utilization pathway (Ellis et al., 1985). Although this promoter has been successfully used to direct the expression of numerous foreign genes, there are circumstances in which it may not be suitable, due to the methanol requirement for induction. In such cases, the strong constitutive promoter from the glyceraldehyde 3-phosphate dehydrogenase gene may be the sole practical alternative (Waterham et al., 1997).
In this paper, we report the isolation and molecular characterization of the PGK1 gene from P. pastoris. Furthermore, we present results on heterologous expression of the α-amylase gene from Bacillus subtilis, which show that the PGK1 promoter may represent an interesting alternative for constitutive heterologous expression in P. pastoris.
Materials and methods
Strains, media and DNA procedures
The P. pastoris strains used in this study were GS115 (his4; Invitrogen) and SMD1168 (his4 pep4). Media for P. pastoris were described in the Pichia Expression kit (Invitrogen): MD (1.34% YNB, 4 × 10−5% biotin, 2% glucose), MDH (MD plus 2 × 10−3% histidine), MDHA (MDH plus 1% starch and 0.08% aspartic and glutamic acid buffer), MGH (1.34% YNB, 4 × 10−5% biotin, 1% glycerol and 2 × 10−3% histidine), MGHA (MGH plus 1% starch and 0.08% aspartic and glutamic acid buffer), MM (1.34% YNB, 4 × 10−5% biotin, 0.5% methanol), MMH (MM plus 2 × 10−3% histidine), MMHA (MMH 1% plus 1% starch and 0.08% aspartic and glutamic acid buffer), YPD (1% yeast extract, 2% peptone and 2% glucose) and YPDS (1% yeast extract, 2% peptone, 2% glucose, 1 M sorbitol).
Escherichia coli strains DH5α (F′ endA1 hsdR17 (rK− mK+) glnV44 thi1 recA1 gyrA (Nalr) relA1 Δ(lacIZYA-argF)U169 deoR (ϕ80dlac Δ(lacZ)M15) and XL1 Blue (F′::Tn10 proA+ B+lacIq Δ(lacZ)M15/recA1 endA1 gyrA96 (Nalr) thi hsdR17(rK− mK+) gln V44 relA1 lac) were routinely used for cloning and plasmid manipulations. These strains were grown in LB medium (0.5% yeast extract, 1% peptone and 1% NaCl) supplied with appropriated antibiotics.
All molecular cloning techniques were carried out as described previously by Sambrook and Russel (2001). Restriction enzymes were obtained from New England Biolabs and Amersham Biosciences and used as detailed by the manufacturer. DNA sequencing analysis was performed on a MegaBace1000 automatic sequencer (Amersham Biosciences).
Cloning of the PGK1 gene by PCR
Based on sequence alignment of the PGK1 gene from the yeasts S. cerevisiae, K. lactis, Y. lipolytica, and Sz. pombe, we designed the degenerated primers C-PGK and PGK-F3 (Table 1), which were used in a PCR reaction consisting of: 10 ng genomic DNA from P. pastoris GS115, 10 pmoles each primer, 2.5 mM MgCl2, 2 U Taq DNA Polymerase (Cenbiot, Brazil) in a final reaction volume of 50 µl. This system was submitted to 30 amplification cycles (30 s/94 °C, 30 s/50 °C and 30 s/72 °C), followed by an elongation cycle (5 min/72 °C). A 689 bp PCR product was cloned into pGEM-T vector (Invitrogen) and the sequence obtained was submitted to Blast analysis.
| Primer | Sequence | RS |
|---|---|---|
| AP | 5′-GGCCACGCGTCGACTAGTAC(T)17 | — |
| AUAP | 5′-GGCCACGCGTCGACTAGTAC | — |
| C-PGK | 5′-AAYAATTCCAAAGAAGCACCAC | — |
| PGK-F3 | 5′-CACTCTTCYATGGTYGG | — |
| PGK-R4 | 5′-GCACCACCCAAGATRGCC | — |
| PGK-FZ | 5′-GAGATCTAGTTGGGTATTCAAATAG | BglII |
| PGK-RZ | 5′-GTTCGAATTTCGTAATCAATTGGGCTATG | BstBI |
| 3PGK-INT | 5′-CGAAAACACCTGGTGGACCGTTCC |
- I, inosine; Y, C/T; R = A/G. Restriction sites (RS) present in the sequence of the primers are underlined.
The 5′ sequences of the PGK1 gene were cloned by PCR using only primer PGK-R4 (Table 1) which anneals within a sequence present in the 689 bp fragment and upstream of it. The reaction conditions were as described above. The 3′-region of the PGK1 gene was obtained through a 3′-RACE experiment (Frohman et al., 1988). Briefly, total RNA from P. pastoris cells grown in MDH was isolated and the cDNA was synthesized using the Super Script Preamplification System Strand cDNA Synthesis kit (Life Technologies), using primer AP (Table 1) following the supplier's recommendations. PGK-F3 and AUAP primers (Table 1) were used to amplify an 800 bp fragment, which was cloned into pGME-T Easy vector (Invitrogen) following DNA sequencing.
Sequence analysis of the P. pastoris PGK1 gene
The nucleotide sequence obtained was analysed through the software programs PHRED (Ewing et al., 1998), PHRAP and CONSED (Gordon et al., 1998). The web interface of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) was used to conduct the search for databank similarity by BLAST search tools. Multiple alignments were performed using the program ClustalW (http://www.ebi.ac.uk/clustalw). TATA-like elements prediction was made using the program HCtata (Hamming-Clustering Method for TATA Signal Prediction in Eukaryotic Genes) (http://125.itba.mi.cnr.it/∼webgene/wwwHC_tata.html). Putative transcription factor motifs were identified using the TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) and MatInspector (http://www.genomatix.de/cgi-bin/matinspector/matinspector.pl) tools.
Southern and Northern Blot analysis
P. pastoris DNA isolation was performed as described by Burke et al. (2000). Approximately 10 µg genomic DNA digested with BamHI, BglII, EcoRI and PvuII were separated on 1% agarose gels and blotted onto nylon membranes. The P. pastoris PGK1 coding sequence, amplified by PCR using primers PGK-F8 and PGK-R9 (1.25 kb), was used as a probe (Table 1). The probe was labelled using the AlkPhos Direct kit (Amersham Biosciences). Hybridization (50 °C) and washing were performed as described by the suppliers. Signal detection was made with the CDP-Star reagent (Amersham Biosciences), according to the manufacturer's instructions.
Total RNA isolation from P. pastoris grown in MDH and MGH media was as described by Stateva et al. (1991). The RNA was analysed by electrophoresis in 1% agarose-formaldehyde gels and transferred to nitrocellulose filters, as described by Sambrook and Russel (2001). The 511 pb DNA probe containing the PGK1 gene fragment from P. pastoris was amplified by PCR with primers PGK-F3 and 3PGK-INT (Table 1), as described for PGK-F3 and C-PGK. Probe labelling, hybridization, washing and signal detection were carried out following the Southern blot protocol. Analyses of RNA abundance were performed using NIH image program and normalized against ribosomal RNA.
Construction of α-amylase expression vectors using PAOX1 and PPGK1
In order to express the B. subtilis α-amylase gene under the control of either the inducible AOX1 promoter (PAOX1) or PPGK1 from P. pastoris, vectors pAOXAMY and pPGKAMY were constructed, respectively (Figure 1). The B. subtilis α-amylase gene lacking the coding sequence for the C-terminal 33 amino acids residues of the enzyme was described previously (Moraes et al., 1995). This truncated α-amylase gene was obtained from the vector pGEM-T/αAMY (Amaral, 2003) by digestion with EcoRI and NotI and inserted into the P. pastoris expression vector pPICZαA (Invitrogen) digested with the same restriction enzymes. The resulting plasmid was named pAOXAMY.

Schematic representation of the strategy used to construct the P. pastoris expression vectors with the PGK1 and AOX1 promoters (PPGK1 and PAOX1, respectively). Vector pAOXAMY was obtained by the cloning of B. subtilis truncated α-amylase reporter gene in the vector pPICZαA, digested with EcoRI–NotI. Vector pPGKAMY was constructed by replacing PAOX1 in pAOXAMY with PPGK1 as follows: pAOXAMY was double-digested with BglII and BstBI and ligated to the ∼2.0 kb PPGK1 fragment, digested with the same enzymes. The resulting vector, pPGKAMYB, was digested with BglII and ligated to the ∼4.0 kb fragment harbouring the HIS4 gene and the 3′AOX1 sequence from pPIC9B, digested with BamHI and BglII, creating plasmid pPGKAMY
Plasmid pPGKAMY was constructed by replacing PAOX1 in pAOXAMY with PPGK1 as follows: pAOXAMY was double-digested with BglII and BstBI, and ligated to a ∼2.0 kbPPGK1 fragment digested with the same enzymes. This promoter fragment was obtained by PCR using primers PGK-FZ and PGK-RZ (Table 1), which were designed to introduce a BstBI site immediately upstream of the ATG translation initiation codon and a BglII site at the 5′ end of the PPGK1 sequence, respectively. The resulting plasmid was named pPGKAMYB. Since PPGK1 did not have suitable restriction sites for plasmid linearization, we decided to add sequences that would allow integration in the AOX1 locus. To do this, pPGKAMYB was digested with BglII and ligated to a ∼4.0 kb DNA fragment harbouring the HIS4 gene and 3′AOX1 sequence from pPIC9B digested with BamHI and BglII, creating plasmid pPGKAMY (pPIC9B was constructed in our laboratory by introducing a BamHI site downstream of the HIS4 gene in pPIC9; Invitrogen). Prior to transformation, plasmids pAOXAMY and pPGKAMY were digested with BglII and BglII/BamHI, respectively, prior to transformation in order to allow their integration at the AOX1 locus.
P. pastoris SMD1168 cells were transformed with the linearized vectors and α-amylase activity was observed through plate assay: pAOXAMY and pPGKAMY transformants were plated either in MDHA, MGHA or MMHA media for 48 h/30 °C and the plates were exposed to iodine vapour to reveal starch hydrolysis halos, as described by Moraes et al. (1995).
Results and discussion
Isolation of the P. pastoris PGK1 gene
In order to clone the PGK1 gene from P. pastoris we used primers based on the sequence alignment of several yeast PGK1 sequences. Using P. pastoris GS115 genomic DNA as template, a PCR with primers PGK-F3 and C-PGK resulted in the amplification of a 694 bp fragment whose sequence showed high identity with the coding region of the PGK1 gene from different yeasts and filamentous fungi. This fragment corresponded to approximately 55% of the predicted PGK1 gene from P. pastoris (Figure 1). A PCR using only PGK-R4 primer amplified a 3.2 kb fragment, which added 495 bp to the coding sequence plus 2.0 kb to the PGK1 5′ upstream regulatory sequences. The 3′ end of the gene was obtained by a 3′ RACE experiment using the PGK-F3 primer, which resulted in the amplification of an 800 bp fragment. The sequence derived from this fragment was confirmed to correspond to the 3′ end of the PGK1 gene by DNA sequencing (Figure 2).

Nucleotide and deduced amino acid sequences of the P. pastoris PGK1 gene. An open reading frame of 1251 bp and the predicted 416 amino acids residues are shown in upper case, while the 5′/3′ non-coding sequences are in lower case. The annealing positions of the primers PGK-F3, PGK-R4 and C-PGK are boxed. On the 5′ non-coding region, a putative TATA box is in bold. Two potential target sequences for Gcr1p are underlined, and the putative heat shock element is double underlined. On the 3′ non-coding region, the putative tripartite element 5′-TAG–TAGT–TTT-3′ is underlined
Sequence analysis of P. pastoris PGK1 gene
Analysis of the approximately 3.3 kb DNA sequences from the P. pastoris PGK1 gene revealed an open reading frame of 1251 bp with the potential to encode a polypeptide of 416 amino acid residues (Figure 2). Although the 3′ end of the gene was derived from cDNA, no introns were observed when the entire PGK1 sequence was obtained by PCR using genomic DNA as template (data not shown). Comparison of the nucleotide and deduced amino acid sequences of the PGK1 gene from P. pastoris and other yeasts showed an overall identity of ∼70% (Table 2).
| Species | P. pastoris | C. maltosa | Y. lipolytica | K. lactis | S. cerevisiae | Sz. pombe |
|---|---|---|---|---|---|---|
| P. pastoris | — | 78 | 76 | 75 | 75 | 72 |
| C. maltosa | 73 | — | 74 | 76 | 74 | 74 |
| Y. lipolytica | 70 | 64 | — | 70 | 70 | 75 |
| K. lactis | 72 | 75 | 65 | — | 82 | 69 |
| S. cerevisiae | 74 | 74 | 66 | 81 | — | 68 |
| Sz. pombe | 66 | 68 | 74 | 66 | 67 | — |
- Identities between predicted amino acid sequences of PGK proteins are shown in the upper part of the table, and identities between PGK nucleotide sequences in the lower. Origin of the sequences: P. pastoris, this work; C. maltosa, GI : 218363; Y. lipolytica, GI : 173251; K. lactis, GI : 2866; S. cerevisiae, GI : 172143; and Sz. pombe, GI : 3184103.
The PGK1 flanking regions presented several features frequently found in other yeast genes. A TATA consensus sequence (TATAAA), which is responsible for correct transcription initiation of the S. cerevisiae PGK1 gene (Rathjen and Mellor, 1990), is present at position −71 relative to the ATG translation initiation codon. Another relevant motif present in the S. cerevisiae PPGK1 is the Gcr1p target sequence. This sequence is essential to achieve high-level activation of glycolytic genes genes in S. cerevisiae (Baker, 1991; Zeng et al., 1997; Deminoff and Santangelo, 2001). Two putative target sequences for Gcr1p, which is composed by three repeats of the CTTCC motif separated by 10 nucleotides from the CAAG motif, were observed in the PPGK1 from P. pastoris (Figure 2). In yeast, several glycolytic genes have their transcription increased in response to heat shock (Mager et al., 1995). In S. cerevisiae the levels of the PGK1 mRNA are enhanced by a 25–38 °C shift in growth conditions (Piper et al., 1986). This increase in mRNA level is mediated by a heat-shock element (HSE) present in the PPGK1 (Piper et al., 1988). In the P. pastoris PGK1 promoter, a putative heat-shock element is also seen at positions −425 to −440 (Figure 2), suggesting that the P. pastoris PGK1 gene is also subjected to thermal regulation. In the downstream sequence, we detected the tripartite element 5′-TAG–TAGT–TTT-3′ (Figure 2) proposed by Zaret and Sherman (1982) as being important for transcription termination in S. cerevisiae. This sequence is observed at the 3′ end of many Y. lipolytica genes, including PGK1 (Dall et al., 1996).
The deduced amino acid sequence alignment of the PGK1 gene from P. pastoris, S. cerevisiae, K. lactis, Y. lipolytica, Sz. pombe and C. maltosa showed highly conserved amino acids (Figure 3). The yeast PGK protein folds into two domains, the N-domain and C-domain, which are composed by the residues 1–185 and 201–394, respectively (McPhillips et al., 1996). The remaining 415 amino acid residues constitute the interdomain linkage (Harlos et al., 1992; Figure 3). Crystallografic studies of porcine PGK confirmed arginines 22, 39, 66, 122 and 170 and histidines 63, 169 and 172 to constitute the binding site for 3-phosphoglycerate in the N-domain (Harlos et al., 1992) and are very similar in several yeast PGK proteins (Figure 3). Conserved residues K219, N336, D374 and T375 form the nucleotide-binding site at the hydrophobic cleft in the C-domain (Fleming and Littlechild, 1997) (Figure 3).
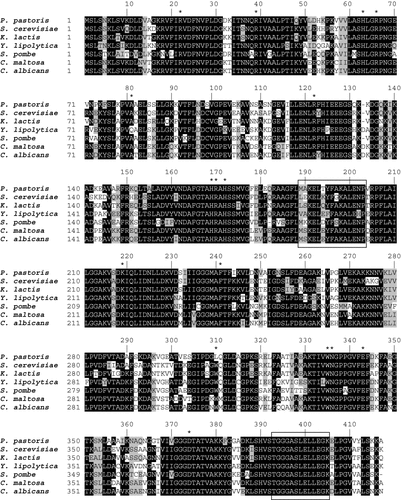
Multiple sequence alignment of PGK proteins from several yeasts. The figure compares amino acids sequences of P. pastoris (this work), C. maltosa (Accession No. D12474 or BAA02040.1), Y. lipolytica (Accession No. M91598 or AAC37504.1), K. lactis (Accession No. X17654 or CAA35646.1), S. cerevisiae (Accession No. J01342 or AAA88729.1) and Sz. pombe (Accession No. GI : 3184103). Black and grey boxes indicate conserved and chemically similar amino acids, respectively. Asterisks indicate important residues for PGK activity. The interdomain amino acids are boxed
Codon usage in P. pastoris
The codon usage (CU) analysis of the P. pastoris PGK1 gene revealed a non-random codon choice, where 11 of the 61 possible amino acid codons are not used (Table 3). The nucleotide of choice in the third position is usually a pyrimidine (61%; Table 3), as was also observed in the PGK1 genes from P. citrinum (Nara et al., 1993) and Trichoderma reesei (Vanhanen et al., 1989). When PGK1 genes from P. pastoris, C. maltosa, Y. lypolitica, K. lactis, S. cerevisiae and Sz. pombe are compared according to CU, they appear very similar (Table 3). The CU of the P. pastoris highly expressed genes PGK1, GAP1 (Waterham et al., 1997) and AOX1 (Koutz et al., 1989) were compared and the biased codon preference stood out (Table 3). Moreover, it was shown that the PGK1, GAP1 and AOX1 genes contain 47%, 50% and 48% G + C, respectively.
| Codon | Amino acid | PGK1 | GAP1 | AOX1 | Cm | Yl | Kl | Sc | Sp | |
|---|---|---|---|---|---|---|---|---|---|---|
| a | b | b | b | b | b | b | b | b | ||
| TTT | phe | 4 | 0.19 | 0.00 | 0.18 | 0.21 | 0.11 | 0.10 | 0.05 | 0.26 |
| TTC | phe | 17 | 0.81 | 1.00 | 0.82 | 0.79 | 0.89 | 0.90 | 0.95 | 0.74 |
| TTA | leu | 5 | 0.12 | 0.00 | 0.04 | 0.33 | 0.00 | 0.00 | 0.12 | 0.00 |
| TTG | leu | 23 | 0.53 | 0.70 | 0.67 | 0.62 | 0.00 | 0.90 | 0.88 | 0.39 |
| CTT | leu | 6 | 0.14 | 0.05 | 0.16 | 0.05 | 0.39 | 0.00 | 0.00 | 0.41 |
| CTC | leu | 1 | 0.02 | 0.10 | 0.04 | 0.00 | 0.44 | 0.00 | 0.00 | 0.20 |
| CTA | leu | 3 | 0.07 | 0.00 | 0.04 | 0.00 | 0.02 | 0.07 | 0.00 | 0.00 |
| CTG | leu | 5 | 0.12 | 0.15 | 0.04 | 0.00 | 0.15 | 0.02 | 0.00 | 0.00 |
| ATT | ile | 7 | 0.35 | 0.43 | 0.42 | 0.47 | 0.39 | 0.43 | 0.39 | 0.21 |
| ATC | ile | 13 | 0.65 | 0.57 | 0.58 | 0.53 | 0.61 | 0.57 | 0.61 | 0.79 |
| ATA | ile | — | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ATG | met | 5 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| GTT | val | 24 | 0.57 | 0.43 | 0.58 | 0.69 | 0.22 | 0.40 | 0.42 | 0.38 |
| GTC | val | 16 | 0.38 | 0.54 | 0.39 | 0.28 | 0.69 | 0.52 | 0.58 | 0.62 |
| GTA | val | 1 | 0.02 | 0.00 | 0.00 | 0.03 | 0.00 | 0.02 | 0.00 | 0.00 |
| GTG | val | 1 | 0.02 | 0.03 | 0.03 | 0.00 | 0.08 | 0.05 | 0.00 | 0.00 |
| TCT | ser | 11 | 0.46 | 0.46 | 0.44 | 0.71 | 0.36 | 0.46 | 0.62 | 0.54 |
| TCC | ser | 10 | 0.42 | 0.50 | 0.42 | 0.24 | 0.59 | 0.33 | 0.23 | 0.42 |
| TCA | ser | 2 | 0.08 | 0.00 | 0.02 | 0.05 | 0.05 | 0.08 | 0.08 | 0.00 |
| TCG | ser | — | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| CCT | pro | 3 | 0.19 | 0.08 | 0.32 | 0.00 | 0.06 | 0.18 | 0.00 | 0.20 |
| CCC | pro | 1 | 0.06 | 0.00 | 0.02 | 0.00 | 0.94 | 0.00 | 0.00 | 0.80 |
| CCA | pro | 12 | 0.75 | 0.92 | 0.66 | 1.00 | 0.00 | 0.82 | 1.00 | 0.00 |
| CCG | pro | — | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ACT | thr | 6 | 0.38 | 0.42 | 0.49 | 0.65 | 0.17 | 0.52 | 0.56 | 0.29 |
| ACC | thr | 6 | 0.38 | 0.54 | 0.43 | 0.35 | 0.83 | 0.43 | 0.44 | 0.71 |
| ACA | thr | 2 | 0.12 | 0.00 | 0.05 | 0.00 | 0.00 | 0.05 | 0.00 | 0.00 |
| ACG | thr | 2 | 0.12 | 0.04 | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| GCT | ala | 27 | 0.57 | 0.71 | 0.58 | 0.80 | 0.34 | 0.67 | 0.74 | 0.61 |
| GCC | ala | 16 | 0.34 | 0.29 | 0.30 | 0.20 | 0.63 | 0.28 | 0.23 | 0.39 |
| GCA | ala | 4 | 0.09 | 0.00 | 0.12 | 0.00 | 0.02 | 0.05 | 0.02 | 0.00 |
| GCG | ala | — | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| TAT | tyr | 1 | 0.25 | 0.00 | 0.00 | 0.2 | 0.2 | 0.00 | 0.00 | 0.00 |
| TAC | tyr | 3 | 0.75 | 1.00 | 1.00 | 0.8 | 0.8 | 1.00 | 1.00 | 1.00 |
| TAA | stop | 1 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| TAG | — | — | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| CAT | his | 1 | 0.17 | 0.00 | 0.09 | 0.00 | 0.25 | 0.33 | 0.13 | 0.00 |
| CAC | his | 5 | 0.83 | 1.00 | 0.91 | 1.00 | 0.75 | 0.67 | 0.87 | 1.00 |
| CAA | gln | 7 | 0.78 | 1.00 | 0.67 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 |
| CAG | gln | 2 | 0.22 | 0.00 | 0.33 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| AAT | asn | 4 | 0.20 | 0.07 | 0.13 | 0.15 | 0.00 | 0.09 | 0.07 | 0.12 |
| AAC | asn | 16 | 0.80 | 0.93 | 0.87 | 0.85 | 1.00 | 0.91 | 0.93 | 0.88 |
| AAA | lys | 9 | 0.22 | 0.08 | 0.13 | 0.89 | 0.00 | 0.13 | 0.05 | 0.00 |
| AAG | lys | 32 | 0.78 | 0.92 | 0.87 | 0.11 | 1.00 | 0.87 | 0.95 | 1.00 |
| GAT | asp | 10 | 0.40 | 0.15 | 0.23 | 0.42 | 0.28 | 0.37 | 0.31 | 0.18 |
| GAC | asp | 15 | 0.60 | 0.85 | 0.77 | 0.58 | 0.72 | 0.63 | 0.69 | 0.82 |
| GAA | glu | 10 | 0.40 | 0.24 | 0.50 | 1.00 | 0.00 | 1.00 | 0.97 | 0.21 |
| GAG | glu | 15 | 0.60 | 0.76 | 0.50 | 0.00 | 1.00 | 0.00 | 0.03 | 0.79 |
| TGT | cys | — | 0.00 | 1.00 | 0.73 | 1.00 | 0.67 | 1.00 | 1.00 | 0.67 |
| TGC | cys | — | 0.00 | 0.00 | 0.27 | 0.00 | 0.33 | 0.00 | 0.00 | 0.33 |
| TGA | — | — | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| TGG | trp | 1 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| CGT | arg | 5 | 0.45 | 0.30 | 0.15 | 0.00 | 0.00 | 0.00 | 0.23 | 0.91 |
| CGC | arg | — | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.09 |
| CGA | arg | — | 0.00 | 0.00 | 0.00 | 0.00 | 0.90 | 0.00 | 0.00 | 0.00 |
| CGG | arg | — | 0.00 | 0.00 | 0.00 | 0.00 | 0.10 | 0.00 | 0.00 | 0.00 |
| AGT | ser | 1 | 0.06 | 0.00 | 0.07 | 0.00 | 0.00 | 0.08 | 0.00 | 0.00 |
| AGC | ser | — | 0.00 | 0.04 | 0.02 | 0.00 | 0.00 | 0.04 | 0.08 | 0.04 |
| AGA | arg | 5 | 0.45 | 0.70 | 0.82 | 1.00 | 0.00 | 1.00 | 0.77 | 0.00 |
| AGG | arg | 1 | 0.09 | 0.00 | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| GGT | gly | 23 | 0.58 | 0.86 | 0.75 | 1.00 | 0.49 | 0.97 | 0.97 | 0.83 |
| GGC | gly | 2 | 0.05 | 0.03 | 0.07 | 0.00 | 0.26 | 0.03 | 0.03 | 0.13 |
| GGA | gly | 15 | 0.38 | 0.10 | 0.19 | 0.00 | 0.26 | 0.00 | 0.00 | 0.05 |
| GGG | gly | — | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
- Fraction of synonymous codon usage in the genes encoding 3-phosphoglycerate kinase from C. maltosa (Cm), Y. lipolytica (Yl), K. lactis (Kl), S. cerevisiae (Sc), Sz. pombe (Sp) and P. pastoris (PGK1); and glyceraldehyde-3-phosphate dehydrogenase and alcohol oxidase 1 from P. pastoris.
- a Total number of specific codons in the P. pastoris PGK1 gene.
- b Fraction of synonymous codon. For example, the amino acid PGK1 gene had 21 Phe codons; of these, 17 were TTC, thus, 17 : 21 = 0.81.
PGK1 gene is represented by a singly copy in P. pastoris genome
In order to determine the number of copies of the PGK1 gene in the P. pastoris genome, a Southern blot experiment was performed (Figure 4). The probe was amplified by PCR and corresponded to the entire PGK1 coding sequence (1.25 kb). A single band was observed with genomic DNA digested with BamHI, BglII and EcoRI, and two bands with genomic DNA digested with PvuII (Figure 4A). The double signal obtained with PvuII-digested DNA can be explained by the presence of an internal site for this restriction enzyme in the coding sequence, which is depicted in Figure 4B. These results strongly suggest that, unlike T. reesei (Vanhanen et al., 1989) the PGK1 gene is represented by a single copy in P. pastoris.
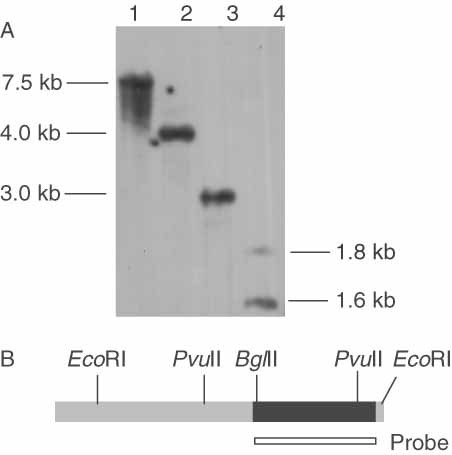
Southern blot analysis of P. pastoris genomic DNA and the PGK1 restriction map. (A) P. pastoris genomic DNA was digested with BamHI (lane 1), BglII (lane 2), EcoRI (lane 3) and PvuII (lane 4). The size of the fragments that hybridized with the probe is shown. (B) The restriction map shows the more relevant sites on the PGK1 gene sequence: 5′/3′ non-conding regions are in grey and the coding sequence (1.25 kb) is represented by a solid black box
Expression pattern of PGK1 gene
In order to examine the regulation of P. pastoris PGK1 gene expression, a Northern blot experiment was carried out. Total RNA from P. pastoris was obtained from glucose- and glycerol-grown cells (Figure 5A) and hybridized against a 511 bp DNA fragment of the P. pastoris PGK1 gene sequence. As shown in Figure 5B, a ∼1.5 kb transcript was detected, based on the relative position of rRNA. This size is in accordance to the predicted size of the mRNA encoded by the PGK1 gene. In S. cerevisiae and A. oryzae, low variation of the mRNA levels was observed when the carbon source was glucose or glycerol (Stanway et al., 1987; Nakajima et al., 2000). The same pattern was observed here, since the P. pastoris PGK mRNA is expressed approximately 2-fold more in glucose- than in glycerol-grown cells (Figure 5B).

Northern blot analysis of the PGK1 expression in two different carbon sources. (A) Total RNA extracted from cells grown in glucose (Glu) and glycerol (Gly). (B) Northern blot analysis using a 511 bp PGK probe. Asterisks represent the relative position of the rRNAs
Analyses of PPGK1 activity by expression of α-amylase gene
To analyse PPGK1 activity, P. pastoris cells transformed with either the pPGKAMY or the pAOXAMY vectors were grown on different carbon sources and the heterologous α-amylase activity assay was performed (Figure 1). As expected, when using glucose or glycerol as carbon sources, the pAOXAMY transformant did not express the B. subtilis α-amylase gene, whereas methanol-grown cells exhibited a starch hydrolysis halo (Figure 6). This occurs due to the repression/induction mechanism of PAOX1 by the carbon source (Tschopp et al., 1987). In glucose- and glycerol-grown cells PAOX1 is fully repressed, while in methanol-grown cells the promoter is maximally induced (Tschopp et al., 1987; Waterham et al., 1997).

Analysis of P. pastoris PPGK1 and PAOX1 promoter activities in directing the expression of the B. subtilis α-amylase gene. P. pastoris SMD1168 cells were transformed with either pPGKAMY (PPGK1) or pAOXAMY (PAOX1) and transformants grown in different carbon sources were analysed for the ability to produce starch hydrolysis halos by iodine vapour staining of starch-containing plates
When analysing expression patterns of glycolytic genes during S. cerevisiae exponential growth in glucose or lactate, several mRNAs showed different levels of induction in glucose, with regulation occurring at the transcription level (Moore et al., 1991). Moreover, in S. cerevisiae the presence of glucose resulted in glycolytic pathway acceleration, decreased respiratory activity, increased ribosome biogenesis and repression of alternative carbon source assimilation pathways (Yin et al., 2000). Therefore, promoter activity analysis of the P. pastoris PGK1 gene in different carbon sources is important for successful heterologous expression when using this promoter. The capability to secrete α-amylase of P. pastoris cells transformed with pPGKAMY, grown in either glucose, glycerol or methanol, was analysed (Figure 6). Cells grown in glucose exhibited the largest hydrolysis halo. This observation is consistent with the Northern blot results (Figure 5B), which showed higher levels of PGK expression in glucose-grown cells. This behaviour was also observed in S. cerevisiae, C. maltosa and A. oryzae, where the highest level of PGK1 gene expression was obtained with glycolytic substrates (Stanway et al., 1987; Masuda et al., 1994; Nakajima et al., 2000). In contrast, in A. nidulans and Y. lipolytica the PGK1 gene shows higher expression levels on gluconeogenic substrates than on glycolytic ones (Streatfield et al., 1992; Dall et al., 1996).
Similarly to the GAP1 promoter (Waterham et al., 1997), PPGK1 from P. pastoris showed high efficiency of expression on the three different carbon sources analysed here. It is worth noting that the size of the α-amylase hydrolysis halos produced by the pPGKAMY transformants were larger than those produced by the pAOXAMY transformants (Figure 6). However, a more detailed promoter strength comparison analysis should be carried out, since the number of copies of the α-amylase gene integrated in the different transformants was not determined here. Nonetheless, based on the preliminary results presented in this work, the PPGK1 promoter isolated in this work may be considered fully functional and constitutive, although its strength varies depending on the carbon source used. Therefore, the utilization of this promoter in the construction of new vectors for constitutive heterologous expression in P. pastoris should be considered.
Acknowledgements
This project was supported by FINATEC and CAPES, Brazil.




