An Orthogonal Covalent Connector System for the Efficient Assembly of Enzyme Cascades on DNA Nanostructures
Abstract
Combining structural DNA nanotechnology with the virtually unlimited variety of enzymes offers unique opportunities for generating novel biocatalytic devices. However, the immobilization of enzymes is still restricted by a lack of efficient covalent coupling techniques. The rational re-engineering of the genetically fusible SNAP-tag linker is reported here. By replacing five amino acids that alter the electrostatic properties of the SNAP_R5 variant, up to 11-fold increased coupling efficiency with benzylguanine-modified oligonucleotides and DNA origami nanostructures (DON) was achieved, resulting in typical occupancy densities of 75%. The novel SNAP_R5 linker can be combined with the equally efficient Halo-based oligonucleotide binding tag (HOB). Since both linkers exhibit neither cross-reactivity nor non-specific binding, they allowed orthogonal assembly of an enzyme cascade consisting of the stereoselective ketoreductase Gre2p and the cofactor-regenerating isocitrate dehydrogenase on DON. The cascade showed approximately 1.6-fold higher activity in a stereoselective cascade reaction than the corresponding free solubilized enzymes. The connector system presented here and the methods used to validate it represent important tools for further development of DON-based multienzyme systems to investigate mechanistic effects of substrate channeling and compartmentalization relevant for exploitation in biosensing and catalysis.
1 Introduction
Scaffolded DNA origami nanostructures (DON) have emerged as powerful and versatile tools for diverse fields, ranging from nanotechnology and materials sciences to biochemistry and biomedicine.[1] Since these self-assembling structures can be used as frameworks for arranging proteins with nanometer precision, a variety of applications are emerging in the field of sensing, biocatalysis, or as tools for studying biological processes.[2] The generation of enzyme-decorated DNA nanoarchitectures is of particular interest with respect to the mimicking of artificial enzyme complexes, as the spatial arrangement of different cooperating catalytically active units can serve to understand molecular mechanisms observed in nature, such as the regulation of activity or substrate channeling,[1, 3] which could be of great benefit for biotechnological applications.[4]
A critical point for the assembly of enzyme-decorated DON concerns the chemical linkage between staple strands of the DNA scaffolds and the enzyme. Ideally, the DNA linkage should be covalent and regiospecific with the enzyme to control the orientation and stoichiometry of the proteins on the origami scaffold. To this end, genetically fusible, self-ligating protein linkers have been used, such as the SNAP- or Halo-tag domains, which covalently bind with a small molecule “suicide ligand” via specific mechanisms (see Figure S1, Supporting Information).[5] While these linkers are superior in terms of coupling yields to other bioorthogonal systems, such as Spy-tag or ybbR,[6] they still do not always result in good occupancy densities of enzymes on the DON.[7] In the case of the Halo-tag, we have recently reported that the efficient self-assembly of protein-decorated DNA structures can be greatly enhanced by fine-tuning the electrostatic interactions between proteins and the negatively charged nucleic acid nanostructures. To this end, by rational engineering of the binding interface of the Halo tag domain, five amino acids located around the entry channel for the chlorohexyl ligand (CH) of the Halo-tag protein were exchanged for positively charged amino acids. The resulting Halo-based oligonucleotide binder (HOB) domain can be genetically fused to any protein-of-interest (POI), and the HOB fusion proteins show significantly improved rates of ligation with CH-modified oligonucleotides and DON.[7] In fact, this variation has improved the reaction of the Halo-tag protein, which is derived from a haloalkane dehalogenase,[8] with CH suicide ligands immobilized on DON to such an extent that occupancy densities of ≈80% are typically achieved.
Another self-ligating protein used for DON modification is the SNAP-tag,[5] a variant of the human O6-alkylguanine-DNA alkyltransferase (hAGT), which specifically reacts with O6-benzylguanine (BG) labeled ligands.[9] However, the SNAP-tag was designed to have no affinity for DNA.[10] This severely limits its use for site-specific immobilization of POIs on DNA nanostructures and thus typically occupancy densities of only 40–60% are achieved, even when a large excess of 1000 equivalents per ligand and longer incubation times are applied.[5] Despite its potential, only very few studies have focused on an improvement of the SNAP-tag for DNA-based applications. For instance, the Morii group has developed a site-specific method to optimize SNAP-tag binding by using a zinc finger protein as a modular adaptor.[11] Although achieving coupling rates of >85%, this method requires the modification of both the POI and the DNA nanostructure with the adaptor and its respective DNA binding domain, respectively. Furthermore, the addition of the zinc finger protein to the SNAP-tag adds additional bulk to the linker and these proteins are sometimes difficult to express in heterologous hosts.
To overcome these limitations, we report here the direct optimization of the SNAP-tag domain to enable SNAP-tagged fusion enzymes that can efficiently bind to DNA nanostructures. Following our previous approach,[7] rational re-design of five amino acids in the SNAP-domain led to an efficient linker for ligation with BG-modified oligonucleotides as well as DNA nanostructures. Furthermore, using a cascade based on a sensitive, stereoselective ketoreductase, and a cofactor-regenerating enzyme, we show that the combination of the new SNAP-tag variant with the HOB-tag provides a powerful orthogonal linkage system to efficiently perform a two-step reduction cascade for the asymmetric synthesis of chiral alcohols on DNA nanostructures.
2 Results and Discussion
2.1 Design of SNAP-Tag Variants
The efficient application of the SNAP-tag as a connector for DNA nanostructures requires a high affinity toward the extremely dense negative charges on the DNA scaffold surface. Based on the previous research devoted to both characterization and optimization of the SNAP-tag, we chose six well-investigated amino acids for mutation to increase affinity toward BG-modified DNA nanostructures (Figure 1A). The mutations K125A, A127T, R128A, S151G, and S152D had previously been introduced in the course of engineering the SNAP-tag for cell biology applications in order to disrupt the protein-DNA interaction exhibited by native hAGT.[10] The residues S151 and S152 were found to interact with the phosphate backbone of double-stranded DNA,[12] while the KAAR motif (K125, A126, A127, R128) plays a crucial role in nuclear retention of hAGT.[13] Based on our experience with the HOB-tag that more efficient connectors for DNA nanostructures become accessible by affecting electrostatic interactions,[7] we hypothesized that reversing A128R and A125K to positively charged amino acids could not only restore overall DNA affinity but also increase coupling efficiencies to the negatively charged DON surface. Moreover, we chose position 160 for mutation, which is located near the active site, and was found to be crucial for affinity toward the BG-ligand.[14] Mutations of amino acid at position 160 modulate substrate specificity, and, depending on the incorporated amino acid, can either increase or disrupt the binding capabilities.[15] Particularly, the exchange of Gly160 with Trp led to a threefold increase in BG-affinity,[9, 16] presumably due to hydrophobic interactions with the substrate, making it an interesting spot for optimized coupling of the SNAP-tag to BG-modified DNA nanostructures.
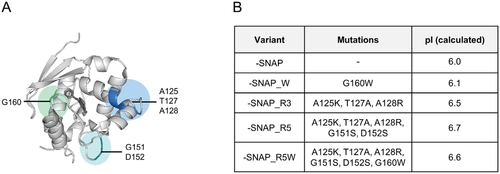
Although the roles of these amino acids in hAGT for DNA affinity have been extensively studied in literature, the research was performed with small-molecule linked substrates and short oligonucleotides.[10, 16, 17] These interaction partners have substantially different properties than DNA nanostructures in terms of negative charge density and steric accessibility. Effects of mutation of these six amino acids have not yet been evaluated for interaction with DNA nanostructures. Therefore, we chose to investigate the above described amino acids regarding their influence on DNA affinity and their use as an optimized immobilization tag, hence generating four different variants, which were compared to the conventional SNAP-tag (Figure 1B). As expected, the variants exhibit an only slightly increased calculated isoelectric point (pI) as compared to the SNAP-tag, since no more than 6 out of 182 amino acids of the protein were changed. However, comparing the electrostatic map of these variants with the original SNAP-tag, the mutant variants bearing the mutations A125K, T127A, A128R, G151S, and D152S show a clear local accumulation of positive charges in the immediate vicinity of the entrance channel (Figure S2, Supporting Information). This suggests a favorable interaction with the negatively charged DNA and is consistent with previous studies in which differences in local charge distribution with only small increases in total pI resulted in increased affinity for DNA.[7]
Furthermore, we also investigated two recently reported alternative tags with respect to their capability for site-specific coupling to DNA nanostructures. The AGT from Sulfolobus solfataricus (SsOGT_wt)[18] as well as an engineered variant SsOGT_H5[19] were described as tags for extremophilic organisms, as they possess a high thermostability. Since they have a specific binding ability to BG substrates comparable to the conventional SNAP-tag, we wanted to test whether they could also be used for protein immobilization in DNA-based applications.
2.2 Coupling Efficacy of SNAP-Tag Variants with Oligonucleotides
The in total seven SNAP variants were genetically fused to the dimeric NADPH-regenerating enzyme ICDH from Bacillus subtilis. The fusion proteins were heterologously expressed in Escherichia coli and purified to homogeneity by Ni-NTA affinity (Figure S3, Supporting Information). For an initial assessment of DNA affinity, we investigated the coupling capabilities of ICDH-SNAP and its variants using a low ratio of 1.3 molar equivalents 5′-BG-modified oligonucleotides per subunit of the dimeric ICDH. The use of a slight excess (1.3 equiv.) of oligonucleotide was chosen to ensure complete consumption of the SNAP protein even at high coupling rates, as established in our previous work.[7] The kinetic analysis revealed significantly altered coupling activity of all rationally designed SNAP variants to the BG-modified oligonucleotide, as compared to the conventional ICDH-SNAP fusion protein (Figure 2A,B; see also Figure S4 for kinetic analysis of all variants; see Figure S5, Supporting Information, for stoichiometric analysis of conjugate formation). In contrast, the thermostable tags SsOGT_wt and SsOGT_H5 showed poor performance (Figure S6, Supporting Information). Therefore, they were considered unsuitable for the application envisaged here and were not used for further investigations. Of the rationally designed SNAP variants, ICDH-SNAP_R5 and -SNAP_R5W performed best and resulted in conjugate formation of up to 95%, with the yield of conjugate exceeding 90% after only 10 min of reaction time.
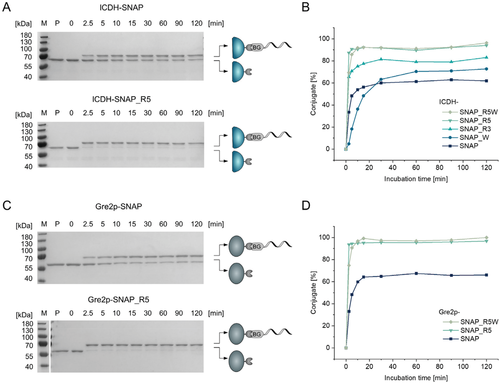
To validate these results and verify applicability of the modified SNAP-tag to other enzymes, the most promising variants R5 and R5W were fused to the monomeric NADPH-dependent enzyme Gre2p from Saccharomyces cerevisiae (Figure S3, Supporting Information), a ketoreductase that catalyzes the stereoselective reduction of prochiral ketone derivatives.[20] As shown in Figures 2C and 2D, almost complete conversion of the 5′-BG-modified oligonucleotide was observed for both Gre2p-SNAP_R5 and -SNAP_R5W, whereas Gre2p-SNAP reached a maximum of only ≈60% conjugate formation. These results thus very clearly confirmed the data obtained with the ICDH variants. The observed increased activity of the R5 and R5W mutants toward BG-modified oligonucleotides is consistent with previous studies,[15, 17] which revealed that mutation of these amino acids plays a critical role in the substrate (BG) and/or DNA binding affinity of the SNAP-tag.
2.3 Immobilization of SNAP-Enzyme Fusions to DNA Origami Nanostructures
Based on the promising results obtained with oligonucleotide conjugation, we then compared the immobilization of ICDH-SNAP_R5, -SNAP_R5W, and conventional ICDH-SNAP on the plane of a quasi 2D rectangular DON that contained three distinguishable BG-modified binding sites (see schematic illustration in Figure 3A,C; for details on the DON design, see Figure S7; for functional analysis of the BG-modified staples see Figure S8, Supporting Information). To enable a direct AFM analysis, a low excess of three molar equivalents of the respective fusion protein per binding site was applied. Binding of conventional ICDH-SNAP led to an average occupancy of only 23% (Figure 3A; see Figure S9, Supporting Information, for large-scale images), whereas ICDH-SNAP_R5 resulted in a much higher occupancy of 74% (Figure 3B). This is a tremendous improvement, as previously occupancies of ≈60% were only achieved with a very large excess of 1000 molar equivalents of the conventional SNAP-tag.[5] The superior binding properties of SNAP_R5 were also observed when only a small excess of 1.3 molar equivalents was used, resulting in occupancy densities of 37% or only 7% for ICDH-SNAP_R5 and ICDH-SNAP, respectively (Figure S10A, Supporting Information). It is also important to note that an enzyme excess of five or more equivalents already limited AFM analysis to such an extent that immobilized proteins could no longer be clearly distinguished (Figure S10B, Supporting Information).
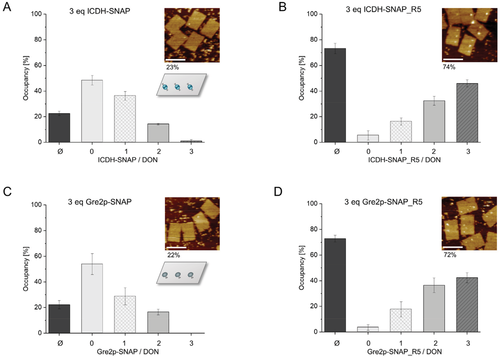
A similar result was obtained with the Gre2p-SNAP_R5 fusion protein, which led to an occupancy density of 72% (Figure 3C), whereas coupling of conventional Gre2p-SNAP resulted in only 22% surface occupancy (Figure 3D). This clearly indicates that the SNAP_R5 mutant is readily applicable to other monomeric enzymes.
Interestingly, a less significant increase in coupling efficiency was observed for ICDH-SNAP_R5W (58%, Figure S11, Supporting Information), indicating that the G160W mutation in this variant increases affinity toward the BG ligand but not toward the negatively charged DNA surface. Indeed, it has been reported that the tryptophan side chain is likely located outside of the BG binding pocket and has a stabilizing effect on the protein-BG complex through stacking interactions with small-molecule BG derivatives.[15] This stabilization could be disrupted by unfavorable interactions between the protein and the bulky DON surface.
To demonstrate the potential of SNAP_R5 for constructing enzyme cascades on DON, the binding efficiency was first compared to that of a co-immobilized HOB-tag fusion enzyme, using the monomeric Gre2p fused to the HOB-tag. To enable AFM analysis as well as accurate determination of enzymatic activity on origami without the interference of unbound enzymes, we employed a bead-assisted purification method of protein-decorated DON,[6] schematically depicted in Figure 4A. In brief, DONs bearing six distinguishable binding sites, three of which equipped with BG- and three with CH-ligands, were additionally equipped with three cleavable biotin linkers (for details, see Figure S7, Supporting Information). These constructs were allowed to bind three molar equivalents of each enzyme per binding site (Gre2p-HOB for CH-ligands and ICDH-SNAP or -SNAP_R5 for BG-ligands) for 120 min and the resulting DON-enzyme constructs were extracted and purified with streptavidin-coated magnetic beads. After cleavage with the reducing agent dithiothreitol (DTT), occupancy densities were determined by AFM analysis. Due to the asymmetric arrangement of the BG- and CH-binding sites on DON, the respective enzyme can be precisely identified and the average distance between the enzymes can be determined (see Figures S7 and S12A, Supporting Information).
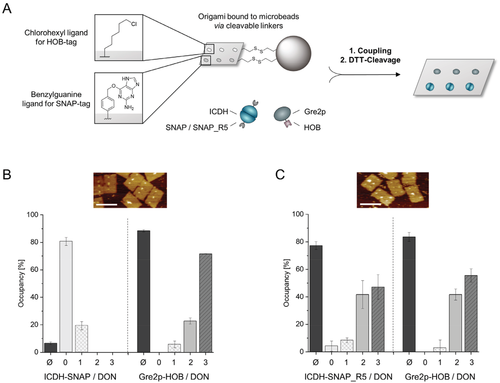
The co-immobilization of ICDH-SNAP with Gre2p-HOB led to a drastically decreased binding of the conventional SNAP-tag of only 7%, while the Gre2p-HOB fusion protein achieved 85% occupancy on the DON. This data translates to a ratio of 1:12.1 of ICDH-SNAP versus Gre2p-HOB (Figure 4B; for large-scale images, see Figure S12B, Supporting Information). In contrast, a remarkably high assembly efficiency of ICDH-SNAP_R5 of 78% was observed, which was similar to the Gre2p-HOB occupancy of 84% and correlates with a ratio of 1:1.1 of ICDH-SNAP_R5 versus Gre2p-HOB (Figure 4C). The improved binding properties of R5 variant were confirmed by electrophoretic analysis of the above coupling reactions (Figure S13, Supporting Information). Hence, these results clearly demonstrate the suitability of SNAP-tag variant R5 for efficient orthogonal coupling of enzymes to DNA nanostructures.
2.4 Construction and Characterization of the Gre2p/ICDH Cascade
Having ensured the efficient construction of the enzyme cascade, we now investigated its functionality. To characterize its catalytic performance, we used the stereoselective reduction of the prochiral substrate 5-nitrononae-2,8-dione 1 (NDK).[20] Gre2p converts NDK 1 with an extraordinary high stereoselectivity, yielding almost exclusively (S)-anti-hydroxy ketone 2 (e.r. > 99:1, Figure 5A). The second reduction of the remaining carbonyl group, which would lead to the corresponding (S,S)-configured diol, is not catalyzed by Gre2p under the usual conditions.[20] Regeneration of the essential cofactor NADPH is achieved by means of co-immobilized ICDH through the oxidation of isocitric acid. Activities of the co-assembled enzyme cascades on DON, non-immobilized (free) enzymes, and negative controls (NC) were quantified by monitoring the formation of 2 using chiral HPLC (Figure 5B). Both controls of free enzymes (1 molar equivalent, 30 nm each), which contained either ICDH-SNAP (blue bars) or ICDH-SNAP_R5 (red bars) in addition to Gre2p-HOB freely present in solution, showed comparable activity. This result thus allowed for direct comparison of the ICDH-SNAP and ICDH-SNAP_R5 cascades and showed that both ICDH variants have comparable catalytic activity.
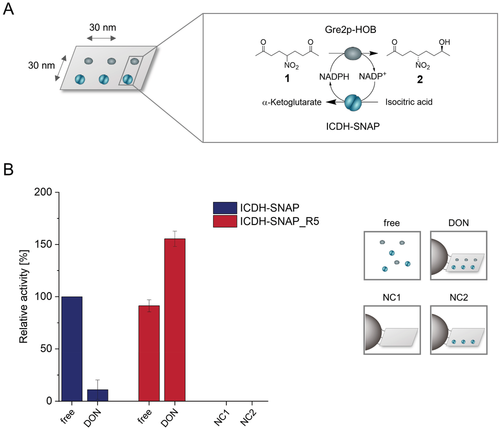
For assembly on DON, three molar equivalents of each enzyme were used per binding site, and the resulting DON-enzyme constructs were bead-purified to eliminate influences from the remaining unbound enzymes. Since DTT, normally used for reductive cleavage of the constructs (see Figure 4A), interferes with HPLC measurements, the biocatalytic cascade reaction was performed directly with the DON coupled on beads. To rule out the possibility that the activity of the enzyme cascade is affected by bead/DON presence, previously performed experiments had shown that comparable activities resulted in the presence or absence of beads and DON (Figure S14, Supporting Information). Furthermore, negative controls were performed to exclude possible nonspecific adsorption of the enzymes to DON or beads. For this purpose, DON equipped either with no binding sites (NC1) or only with BG binding sites (NC2) were immobilized on the beads and incubated with the mixture of Gre2p-HOB and ICDH-SNAP_R5 and bead-purified. NC2, in which only ICDH-SNAP_R5 was bound on the beads, also served as a control for the fact that for a successful cascade reaction, both enzymes must indeed be present and co-immobilized to yield the product. As expected, no formation of the hydroxy ketone 2 product was observed in either control (Figure 5B), confirming that all unbound enzymes were successfully removed by bead-purification.
Directional assembly of Gre2p-HOB and ICDH on DON revealed remarkable differences in activities between the ICDH-SNAP- and ICDH-SNAP_R5-based cascades. When the conventional SNAP-tag was used in the cascade mounted on DON (blue), a much lower product formation of 2 was observed, which can thus be attributed to the poorer binding properties and the resulting removal of ICDH-SNAP by washing steps in the bead cleaning process. In contrast, the cascade with ICDH-SNAP_R5 resulted in the efficient formation of 2, confirming that this SNAP-tag variant allows for efficient assembly of the enzyme cascade on DON. Interestingly, the directional assembly of Gre2p-HOB and ICDH-SNAP_R5 on DON resulted in ≈60% higher overall activity compared to the enzymes in free solution. To corroborate this result, the enzyme amounts in the samples of free and immobilized Gre2p-HOB and ICDH-SNAP_R5 were additionally quantified by Western blot analysis (Figure S15, Supporting Information), and the results confirmed that the observed increase in activity was indeed due to the directional nanoscale assembly and not to different enzyme amounts.
The here observed increase in catalytic activity of enzyme cascades on DNA scaffolds has previously been observed in several studies. For instance, Hao Yan and coworkers observed an about twofold increase in a DON assembled Glucose Oxidase (GOx) horseradish peroxidase (HRP) cascade, where the enzymes were positioned in distances between 10–45 nm, and this effect was attributed to distance-dependent substrate diffusion with the help of modeling.[21] However, Hess and coworkers have shown that proximity alone does not contribute to activity enhancement in the GOx-HRP cascade,[22] which led them to emphasize that substrates in nature are channeled by confinement rather than proximity[23] and to propose design principles for a compartmentalized enzyme cascade reaction.[24] We therefore hypothesize that the enhanced cascade activity observed in our study is likely due to effects such as diffusion constraints in compartmentalized microenvironments generated by the DNA nanostructures on the microbeads. Further detailed studies as well as refined methodological approaches are needed to clarify the mechanistic origin of these phenomena and to exploit them for technical applications.[1, 4, 23, 25] It seems feasible that the efficient connector system, in combination with robust controls, the bead-based purification and quantifications by AFM and Western blot analysis shown here, point in the right direction to generate the quantitative data needed for this purpose.
3 Conclusion
In summary, by re-engineering five amino acids of the established SNAP-tag, we created an effective, genetically fusible connector for site-selective immobilization of enzymes on DNA-based nanostructures. The resulting SNAP_R5 fusion proteins showed up to 11-fold increased coupling efficiency toward BG-modified oligonucleotides as well as DON as compared to the conventional SNAP-tag, leading to typical occupancy densities of 75%. By combining the new SNAP linker with the equally efficient HOB-tag, orthogonal coupling of the sensitive, stereoselective ketoreductase Gre2p and the NADPH-regenerating ICDH on DON was enabled. Since both linkers exhibit neither cross-reactivity nor non-specific binding, they allowed orthogonal assembly of an enzyme cascade that exhibited an activity approximately 1.6-fold higher than that of the corresponding biocatalytic reaction with free solubilized enzymes. Although this result is quantitatively consistent with some other cascades described previously, there is agreement that further detailed studies using robust methodological approaches and tools are needed to elucidate the mechanistic origin of the underlying effects and exploit them for technical applications.[1, 4, 23, 25] We believe that the efficient connector system presented here and the methods used for its validation represent an important contribution to the further development of DON-based multienzyme systems.
Acknowledgements
This work was supported through DFG Project Ni 399/15-1 and the Helmholtz program “Materials Systems Engineering” under the topic “Adaptive and Bioinstructive Materials Systems”. The authors thank Camilla Stolle for experimental help.
Open access funding enabled and organized by Projekt DEAL.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.