Synthetic community improves crop performance and alters rhizosphere microbial communities
Abstract
Introduction
Harnessing synthetic communities (SynCom) of plant growth-promoting (PGP) microorganisms is considered a promising approach to improve crop fitness and productivity. However, biotic mechanisms that underpin improved plant performance and the effects of delivery mode of synthetic community are poorly understood. These are critical knowledge gaps that constrain field efficacy of SynCom and hence large-scale adoption by the farming community.
Material & Methods
In this study, a SynCom of four PGP microbial species was constructed and applied to either as seed dressing (treatment T1, applied at the time of sowing) or to soil (treatment T2, applied in soil at true leaf stage) across five different cotton (Gossypium hirsutum) cultivars. The impact of SynCom on plant growth, rhizosphere microbiome and soil nutrient availability, and how this was modified by plant variety and mode of applications, was assessed.
Results
Results showed that the seed application of SynCom had the strongest positive impact on overall plant fitness, resulting in higher germination (14.3%), increased plant height (7.4%) and shoot biomass (5.4%). A significant increase in the number of flowers (10.4%) and yield (8.5%) was also observed in T1. The soil nitrate availability was enhanced by 28% and 55% under T1 and T2, respectively. Results further suggested that SynCom applications triggered enrichment of members from bacterial phyla Actinobacteria, Firmicutes and Cyanobacteria in the rhizosphere. A shift in fungal communities was also observed, with a significant increase in the relative abundance of fungi from phyla Chytridiomycota and Basidiomycota in SynCom treatments. A structural equation model suggested that SynCom directly increased crop productivity but also indirectly via impacting the alpha diversity of bacteria.
Conclusion
Overall, this study provides mechanistic evidence that SynCom applications can shift rhizosphere microbial communities and improve soil fertility, plant growth, and crop productivity, suggesting that their use could contribute toward sustainable increase in farm productivity.
1 INTRODUCTION
Crop performance is strongly driven by biotic and abiotic environmental factors. For example, microbial pathogens can reduce plant fitness and productivity significantly. Likewise, the absence of key beneficial microbes can limit the bioavailability of key nutrients including nitrogen (N), phosphorus (P), and sulphur (S) which are essential for plant growth and productivity (de Souza et al., 2020; Oldroyd & Dixon, 2014; Singh & Trivedi, 2017). Harnessing beneficial microbes that promote plant fitness by increasing nutrient supply and use efficiencies is fast emerging as a complementary approach to traditional use of chemical fertilisers (Singh & Trivedi, 2017; Tabassum et al., 2017). Plant growth-promoting rhizobacteria (PGPR) applications can exert multiple plant-beneficial functions such as phytohormone provision, nutrient solubilisation (Mukherjee et al., 2020; Trivedi et al., 2011). PGPR-mediated plant performance and fitness often result in improved resistance to abiotic and biotic stresses (Batista & Singh, 2021; Mukherjee et al., 2020; Qiu et al., 2019; Trivedi et al., 2011).
Despite potential benefits, the adoption of PGPR tools in agriculture remains below expectation mainly due to inconsistent field efficacy (Batista & Singh, 2021; Qiu et al., 2019). In many cases, microbial inoculant products that contain single species either fail to colonise plant in field conditions or do not provide expected benefits (Dakora, 2003; Korir et al., 2017; Qiu et al., 2019). Conversely, it is increasingly recognised that the use of synthetic microbial communities (SynComs) can overcome some of these challenges (Trivedi et al., 2021). SynComs include multiple microbial species which can grow together and provide collective benefits to the plant host. Synergistic interactions between compatible but functionally diverse microbial strains can lead to better adaptation to new environments and simultaneously offer a broader range of plant growth-promoting functional traits to improve plant phenotypes and ultimately enhance crop productivity (Hays et al., 2015; Parnell et al., 2016; Pereg & McMillan, 2015; Singh & Trivedi, 2017). However, our understanding of the mechanisms that underpin these positive impacts on plant fitness remains poor, constraining the development and adoption of effective SynCom tools. Indeed, SynComs can affect plant performance directly by providing nutrients, phytohormones, and/or resistance against pathogens, or indirectly, by altering soil physicochemical status/modifying physiology and immune response of host plant, and composition of plant microbiome (Pereg & McMillan, 2015; Trivedi et al., 2021). For example, recent studies in pepper and tomato have highlighted positive impacts of PGPR on rhizosphere microbiome assembly and richness indices, with increases in the abundance of key plants beneficial microbes (Zhang et al., 2019). It is also reported that different plant genotypes harbour different microbiomes, however, it is not known how genotype-linked plant microbes will respond to introduced SynCom inoculants and consequences for host functions (Hamonts et al., 2018; Qiao et al., 2017). However, the relative contribution of different processes (e.g., nutrient provision and recruitment of beneficial microbial taxa) that collectively underpin SynCom effects is poorly understood.
Timing and mode of application can also have impacts on microbial inoculant efficacy. For example, an early stage inoculation such as seed dressing could provide an advantage (i.e., priority effect) for beneficial microbial colonisation in the rhizosphere and root endosphere (Qiu et al., 2019). On the other hand, direct soil application has a logistic advantage as it can be integrated in irrigation or fertilisation systems, although, in late application, effective colonisation of plant roots by inoculants may be constrained by competition from indigenous microflora (Y. Liu et al., 2021; Qiu et al., 2019) However, experimental evidence on the impact of timing and mode of delivery on the efficacy of inoculants remain an important knowledge gap which constrains our ability to develop the best possible application approach (Backer et al., 2018; Trivedi et al., 2021).
Cotton (Gossypium hirsutum) is a major cash crop of the world (USDA, FAS, 2018) that requires high fertiliser inputs and irrigation to maintain productivity. However, excessive use of fertilisers can have significant negative consequences for environmental sustainability, particularly water pollution and nitrous oxide emissions (Berg, 2009; Pereg & McMillan, 2015). There is an increasing demand for a sustainable farming practice that reduces the usages of agrochemicals while maintaining/increasing farm productivity (Basu et al., 2021; Batista & Singh, 2021; Pereg & McMillan, 2015). Microbial-based biological solutions, especially in form of SynComs, are considered among the most promising approach to address these challenges but experimental evidence remains limited (Basu et al., 2021; Trivedi et al., 2021; Uzoh & Babalola, 2018).
In this study, we aimed to evaluate the ability of a bacterial SynCom (i.e., a combination of bacteria harbouring different plant-beneficial and synergistic traits) to promote cotton growth and productivity and to assess the SynCom effect on rhizosphere microbial community structure. Additionally, the effect of inoculation mode was assessed by comparing seed-dressing (i.e., SynCom application at the time of sowing) and soil treatment (i.e., SynCom application at 4-week growth stage) on plant yield-related parameters. We hypothesised that: (1) SynCom application would enhance plant growth and yield, with seed-dressing application having a stronger impact on yield compared to soil application due to priority effect; and (2) the impact of SynCom application on crop performance will be partially explained by a shift in rhizosphere microbial structure.
2 MATERIAL AND METHODS
2.1 Experiment setup, SynCom-treatment preparation, and delivery
Four PGPR (Table 1) were carefully selected from a laboratory library of microbes isolated from a cotton farm, based on their PGP traits (i.e., indole acetic acid [IAA0] production, phosphorus [P0]-solubilisation, ammonium [NH3] production, and Fusarium oxysporum f. sp. vasinfectum biocontrol properties; Table 1) and in planta growth-promoting effects. Before preparation of the SymCom treatment, the compatibility of the microbial candidates was confirmed using a synergistic assay following the method from a previous study (Berendsen et al., 2018), whereby the isolates were considered compatible when no growth inhibition was observed when grown on the same plate. For the inoculum preparation, each isolate was revived from the glycerol stocks on NA (Nutrient Agar) and transferred separately into 20 ml of nutrient broth by loop transfer then incubated at 28°C for 48 h at 100 rpm (RATEK, Rowe Scientific Pty Ltd.). Bacterial cells were harvested by centrifugation at 4000g for 10 min, then resuspended in 20 ml sterilised saline water (0.8% NaCl).
| SynCom PGPR candidates | PGP traits | FOV inhibition | Compatibility | Biofilm formation | Accession no. NCBI |
|---|---|---|---|---|---|
| Arthhrobacter sp. | IAA | + | + | + | MH680887 |
| Enterobacter sp. | IAA, PS, Ammonia | + | + | + | MT158576 |
| Brevibacterium sp. | Ammonia | + | + | + | MH680885 |
| Plantibacter sp. | PS | + | + | + | MH680891 |
- Abbreviations: FOV, field ofview; PGPR, plant growth-promoting rhizobacteria.
A pot experiment was set up at Western Sydney University, Richmond Australia (33°36'37.3"S 150°44'48.9"E) to examine the impacts of SynCom on plant growth, yield, soil nutrient availability and rhizosphere microbial communities. Three treatments were applied to five cotton cultivars in triplicates, and consisted of: (1) C-uninoculated control (2) T1-SynCom seed dressing at time of sowing, and (3) T2-SynCom-soil treatment at true leaf stage. Five cotton cultivars encompassing the most widely cultivated varieties of Australia were used in this study and included V1-CIM448, V2-Sikora, V3-CS50, V4-DP16, V5-Sicot BRF71 to examine cultivar-dependent response to SynCom treatments. Briefly, 60 seeds per cultivar were sterilised with 70% ethanol for 30 s and rinsed with sterile distilled water, followed by a 3% sodium hypochlorite solution treatment for 3 min, with regular shaking by hand (Yao et al., 2010). Seeds were then thoroughly washed with sterilised distilled water. For the SynCom seed dressing application (T1), seeds were soaked for 2 h in 20 ml of SynCom suspension (consisting of equal concentration of four bacteria, with an adjusted concentration of 108 CFU per ml by checking OD at 600 nm on Nanodrop™), whereas control seeds were soaked in sterilised saline water.
The experiment was conducted during the cotton growing season for 24 weeks (October 2018–April 2019) under outdoor conditions. Sandy loam soil (pH 6.9) was collected (0–10 cm) from an adjacent field that had a history of canola, cotton, and vegetable cultivation. Each pot was 15 cm in diameter and 40 cm deep and pots were kept 20 cm apart from each other. A common fertiliser (Yates “Thrive Soluble All Purpose Plant Food” with NPK ratio-25:5:8/kg) was applied at a rate of 80 kg/ha. Yates Lime Sulphur (200 g/L Sulphur) was applied at monthly intervals to control for mites/other pests as per manufacturer instructions. Before seed sowing, pots were irrigated to 60% field capacity and then daily manual watering was performed. Watering was withheld during rainy days. Seed sowing was performed by adding six seeds per pot at a depth of 3 cm, with an additional 5 ml of SynCom suspension added in T1 pots, while the control and T2 pots received the same amount of sterilised water.
Thinning was performed at the 12th day after sowing (DAS) with three plants maintained in each pot. Treatment T2-SynCom soil treatment was applied at the true-leaf stage (TL-stage; leaves developed from the cotyledons or up to 4-week stage), whereby the soil of each pot was drenched with 25 ml of the SynCom suspension (108 CFU/ml adjusted by following the same protocol as seed dressing treatments was prepared) (Jetiyanon & Kloepper, 2002; Kumar & Gera, 2014; Myresiotis et al., 2012; Niknam & Dhawan, 2003), while the control and T1 pots received the same amount of sterilised saline water.
2.2 Plant measures and soil nutrient parameters
Germination rate was recorded until the 12th DAS. Plant height (up to tip of main stem), root length at TL (4-week) and harvest stage (24-week) and fresh/dry biomass were measured at the TL stage, a number of flowers were counted, and cotton bolls were picked on maturation (when buds were fully opened) and seed cotton (lint + seed) weight was recorded as a measure of crop productivity.
For soil analyses, rhizosphere soil samples were collected at TL and flowering stages. At each sampling point, one cotton plant from each pot was carefully uprooted from the soil. Soil tightly attached to the roots (i.e., rhizosphere soil) was collected for DNA extraction in 2 ml sterilised microcentrifuge tubes and immediately stored at −20°C until further analysis. Extractable nutrients ammonia [NH4+], nitrate [NO3−] and phosphorus [PO43−] in bulk soil samples were also determined at TL (4-week) and flowering (12th week) stage. These stages were considered as the most actively growing stages. Ammonia [NH4+] and NO3− were measured following extraction from fresh soil with 2 M KCl (Keeney & Nelson, 1982) and determined on SEAL AQ2 (SEAL Analytical). Extractable soil phosphorus (P) was determined spectrophometrically (880 nm) following extraction in 0.5 M sodium bicarbonate (Olsen et al., 1954) and PO4 was determined on SEAL AQ2 Analyzer following acidification with 12 N sulphuric acid.
2.3 DNA extraction from rhizosphere soils
Total genomic DNA was extracted from ~200 mg of rhizosphere soil using DNeasy PowerSoil Pro Kit (Qiagen), following the manufacturer's instructions. Extracted DNA was quality checked by NanoDrop 2000 (Thermo Fisher Scientific), and PCR checked to confirm the amplifiability. Amplicon sequencing was performed using 799F/1193R targeting 16S rRNA for bacteria and ITS2 region FITS7-ITS4R (Hamonts et al., 2018; H. Liu et al., 2019; Qiu et al., 2020) at the Next-Generation Sequencing Facility, Western Sydney University, Richmond, NSW Australia.
2.4 Microbial community amplicon sequence analysis
Raw sequence data obtained were processed using Mothur standard operating procedure (Schloss et al., 2009). Briefly, forward and reverse sequences were merged into contigs. Sequences that contained unidentified bases or had greater than eight homopolymers were filtered out. Bacterial sequences were aligned against Silva 16 S rRNA gene database version 132 (Pruesse et al., 2007). Aligned bacterial sequences and unaligned fungal sequences were pre-clustered at cutoff of diffs = 1 and 2, respectively, before chimera were identified and removed using UCHIME (Edgar et al., 2011). Additionally, singleton was removed to reduce read error (Reeder & Knight, 2009). Bacterial and fungal sequences were then taxonomically classified according to the Silva database version 132 and UNITE database version 8, respectively, with 60% cutoff confidence and sequences that match cotton mitochondria, chloroplast, archaea (bacteria), and host ITS regions were removed. The remaining sequences were clustered into Zero-radius Operational Taxonomic Units (ZOTUs) at 100% identity and taxonomy was assigned.
2.5 Microbial community structure
Microbial community analysis was carried out at two stages: (1) TL-stage where we compared T1 and control treatment; and (2) flowering stage, where we compared T1, T2, and control treatments. Bacterial and fungal OTU richness, Chao1, and Shannon indices were calculated with “Phyloseq” R package. Pairwise Bray-Curtis dissimilarity metrics of square-root-transformed bacterial/fungal community structure were calculated, and differences of bacterial and fungal community composition among cultivars and treatments were estimated using a PERMANOVA with 9999 permutations, using the “adonis” function in the R package “vegan” (Oksanen et al., 2012). Bacterial and fungal community structure were visualised using principal coordinate analysis (PCoA) based on the OTU feature tables using R packages “phyloseq” and “ggplot2” (McMurdie & Holmes, 2013). To identify the characteristics OTUs that were enriched by SynCom treatment, an indicator analysis combining both the abundance and occurrence of a given OTU across all treatments was used. The indicator values (IndVal)of each OTU were calculated using the “multipatt” function in the R package “indicspecies,” with i = 999 random permutations (Cáceres & Legendre, 2009; De Cáceres et al., 2010).
2.6 Statistical analyses
Two-way analysis of variance (ANOVA) and Tukey HSD test were used to evaluate the effect of treatments (cultivar and SynCom application) on plant and soil parameters at two stages: (1) TL-stage where we compared T1 and control treatment; and (2) flowering stage, where we compared T1, T2, and control. Data that did not meet the assumptions of the ANOVA were transformed before statistical analysis. Treatment-wise means were plotted as box plots and compared the overall effect of SynCom vs Control. For all statistical analyses, treatment effects were determined to be statistically different at p < 0.05. Two-way ANOVA and Tukey HSD test analysis were conducted by using “Agricolae” R package.
A structural equation model (SEM) was built to determine the casual pathways through which SynCom application influenced the crop productivity, where both direct and indirect (via changing microbial community and, nutrient availability) pathways were considered. The initial model also included cultivar identity as a controlling factor. We simplified the initial model by eliminating nonsignificant pathways and state variables based on regression weight estimates. Shannon index of bacterial and fungal communities was used as a measure of alpha diversity. We also performed principal coordinates analysis (PCoA) in the R package vegan (R version 4.0.2) (Dixon, 2003) based on the Bray–Curtis dissimilarities of square-root-transformed bacterial community operational taxonomic units (OTU) composition and PCoA scores of the first axis were used as proxies for changes in community composition in the SEM analysis. Before conducting SEM, soil phosphate and [NO3−]/[NH4+] ratio was square-root-transformed to improve linearity. The model was then parameterised and its overall goodness of fit tested using the Chi-square test, the Comparative Fit Index (CFI), the Root Mean Square Error of approximation (RMSEA), and the (standardised) Root Mean Square Residual (SRMR) (Kline, 2012). All SEM analyses were conducted using the R package “lavaan” (Rosseel, 2012).
3 RESULTS
3.1 Impact of SynCom seed dressing (T1) on germination and early plant performance
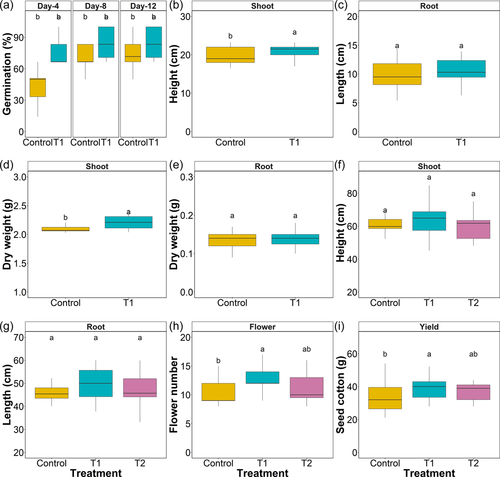
Germination rate was significantly (14.3%) higher in T1-SynCom seed dressing compared to control treatments at 12th DAS (p < 0.05) (Figure 1a, Table S1). Plant shoot height was significantly (7.4%) higher in T1 plants than control (p < 0.05). Cultivars and treatment x cultivar also had a significant effect (p < 0.01 and 0.05, respectively) on plant height (Figure 1b, Tables S1 and S2). Root length exhibited significant effect of the cultivar and treatment × cultivar (p < 0.001). T1 treatment increased root length by 9.27% compared with control plants although differences were marginally insignificant (p = 0.053) (Figure 1c, Tables S1 and S2). For plant biomass, shoot dry weight was significantly (5.14%) higher (p < 0.01) in T1 compared with control plants (Figure 1d, Table S1). However, root dry weight did not change after SynCom application, whereas cultivar exhibited a significant effect (p < 0.001) (Figure 1e; Tables S1 and S2).
3.2 Impact of T1 and T2 treatments on plant growth at harvest stage
Plant shoot height (Figure 1f, Table S3) measured at the harvest stage was not affected by treatment or cultivar (p = 0.335 and 0.064, respectively), although interaction of treatment × cultivar was significant (p < 0.05). Root length (Figure 1g, Table S3 and S4) exhibited significant effect of treatment × cultivar interaction (p < 0.001). However, no effects were observed for cultivar and treatments individually. Flower number per plant (Figure 1f, Table S3 and S4) was positively influenced by SynCom treatments (p < 0.01), where T1 elicited 10.44% and 7.36% higher flowers per plant compared to control and T2-SynCom soil treatment, respectively. For the plant productivity (Figure 1i, Table S3 and S4) measures taken as a combined weight of seed and lint, a significant effect of treatment (p < 0.05) and cultivar (p < 0.001) was observed. Among treatments, T1 and T2 showed 8.55% and 5.3% higher yield over control (p < 0.018 and 0.193, respectively). There was no significant interaction between treatment and cultivar suggesting that SynCom treatments have similar effect on productivity across all cultivars.
3.3 Soil ammonium [NH4+], nitrate [NO3−] and phosphorus [PO43−] availability at TL stage and flowering stage
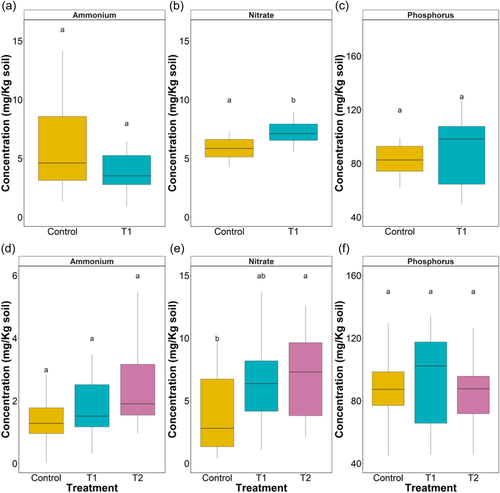
At the TL-stage, soil NH4+ concentrations (Figure 2a, Table S1) were not significantly different between treatments or cultivar (p > 0.05). In general, 40.5% higher ammonium concentration was recorded in control over T1 soil samples (C = 6.0 ± 1.0 SE and T1 = 4.2 ± 0.6 mg/Kg). The interaction effect of treatment × cultivar was also significant (p < 0.05). Soil extractable NO3− was 28.8% higher in T1 compared with control (p < 0.01) (Figure 2b, Table S1). Soil available P (Figures 2c, S1, Table S1) data suggested no independent impact of treatment and cultivar. A significant interaction between cultivar and T1 treatment (p < 0.05) was observed, with cultivar V1 and V4 showing higher soil available P than their controls.
At flowering stage, soil NH4+ concentrations were not significantly impacted by cultivars or treatments. Conversely, soil nitrate availability (Figure 2e, Table S3) was significantly different between treatments (p < 0.01) and cultivars (p < 0.05). Overall, NO3− was 55% higher in T2 compared with control soils (p < 0.05). Soil available P concentration was not affected by treatment or treatment × cultivars (Figure 2f, Table S3).
3.4 Microbial diversity
At TL stage, no significant difference (p > 0.05) was observed in bacterial OTU richness between treatments (Figure 3a, Table S1). However, the interaction impact of treatment × cultivar was significant (p < 0.05). Fungal OTU richness showed no difference across treatments or cultivars (Figure 3b, Table S1). A higher value of bacterial Chao1 index in control versus T1 and a significant effect of cultivars and treatments independently (p < 0.05 and p < 0.001, respectively; Table S1) were observed. No trend was recorded in Chao1 index of fungal community (Figure 3d). Shannon index of bacteria changed by treatments where lower Shannon diversity was found in T1 compared with control (p < 0.05). The Shannon index of fungi did not respond to either treatment or cultivars (Figure S2a,b, Table S1).

At flowering stage, bacterial richness (Figure 3e, Table S3) did not differ between treatments, however fungal richness (Figure 3f, Table S2) showed strong treatment (p < 0.001), cultivar (p < 0.01), and treatment × cultivar effects (p < 0.001). For bacteria, the Chao1 index (Figure 3g, Table S2) showed significant treatment effect (p < 0.001), and both T1 and T2 had higher richness than control, whereas no effect of cultivar and treatment × cultivar was observed. Similarly, the fungal Chao1 index (Figure 3h, Table S3) was higher in T1 and T2 treatments than control soil samples (p < 0.05). The fungal Chao1 index was significantly impacted by treatment (p < 0.001), cultivar (p < 0.001) and treatment × cultivar (p < 0.001). Shannon index of bacteria and fungi were lower for T1 and T2 compared to the control treatment (Figure S2c,d). For bacteria, the Shannon Index was significantly affected by treatment and interaction of treatment × cultivar (p < 0.001 and 0.01, Table S3), whereas fungi Shannon index exhibited significant effects of treatment, cultivar, and treatment × cultivar (p < 0.001, 0.05, and 0.001) (Table S3).
3.5 Community structure of bacterial and fungal communities
The PCoA analyses using Bray–Curtis distance matrices showed that bacterial communities were separated according to treatment on the first two coordinate axes that explain 46.7% and 20.7% of the variation, respectively (Figure S3a). This was further confirmed by PERMANOVA where significant differences in communities were observed due to treatment (p < 0.05, Table 2) and no effect observed due to cultivars (p > 0.05). Similarly, PCoA analysis of fungal communities at TL stage (Figure S3b) showed separation of treatments at the first two coordinate axes (axis 1: 13.4% and axis 2: 11%). PERMANOVA showed significant effect of treatments (p < 0.05, Table 2). PCoA of bacterial at flowering stage (Figure S4a) showed that there was no significant difference between treatments and cultivar effects. In fungal communities at flowering stage, a clear separation of T2 from controls was observed (Figure S4b). Further PERMANOVA indicated that treatment and cultivars were significantly impacting on fungal community structure (p < 0.001 for all, and Table 2).
| df | SumsOfSqs | MeanSqs | F.Model | R2 | Pr(>F) | |
|---|---|---|---|---|---|---|
| Bacteria comparison 1: Control and T1 (True-leaf and flowering stage) | ||||||
| Treatment | 1 | 0.734 | 0.7348 | 3.749 | 0.053 | 0.035 |
| Cultivar | 4 | 0.563 | 0.140 | 0.719 | 0.041 | 0.819 |
| Treatment:Cultivar | 4 | 0.937 | 0.234 | 1.196 | 0.068 | 0.419 |
| Residuals | 50 | 11.505 | 0.230 | 0.837 | ||
| Total | 59 | 13.740 | 1.000 | |||
| Bacteria comparison 2: Control, T1 & T2 (Flowering stage) | ||||||
| Treatment | 2 | 0.7907 | 0.39537 | 1.79217 | 0.07963 | 0.129 |
| Cultivar | 4 | 0.6751 | 0.16878 | 0.76505 | 0.06799 | 0.629 |
| Treatment:Cultivar | 8 | 1.8455 | 0.23069 | 1.04571 | 0.18586 | 0.4195 |
| Residuals | 30 | 6.6182 | 0.22061 | 0.66652 | ||
| Total | 44 | 9.9296 | 1 | |||
| Fungi comparison 1: Control & T1 (True-leaf and Flowering stage) | ||||||
| Treatment | 1 | 0.425 | 0.425 | 1.530 | 0.025 | 0.039 |
| Cultivar | 4 | 1.152 | 0.288 | 1.035 | 0.069 | 0.350 |
| Treatment:Cultivar | 4 | 1.162 | 0.290 | 1.050 | 0.069 | 0.327 |
| Residuals | 50 | 11.505 | 0.230 | 0.835 | ||
| Total | 59 | 13.740 | 1.000 | |||
| Fungi comparison 2: Control, T1, & T2 (Flowering stage) | ||||||
| Treatment | 2 | 2.2325 | 1.11623 | 6.6303 | 0.18465 | 1.00E−04 |
| Cultivar | 4 | 1.6905 | 0.42263 | 2.5104 | 0.13983 | 1.00E−04 |
| Treatment:Cultivar | 8 | 3.1164 | 0.38954 | 2.3139 | 0.25777 | 1.00E−04 |
| Residuals | 30 | 5.0506 | 0.16835 | 0.41775 | ||
| Total | 44 | 12.0899 | 1 | |||
3.6 Indicator species analysis
Indicator species analysis revealed that at TL-stage for bacteria, OTUs from phyla Proteobacteria (genera Betaproteobacteriales_unclassified, Gammaproteobacteria_unclassified, Beijerinckiaceae_unclassified, Azospirillaceae_unclassified, Acidibacter, Rhizobiales_unclassified, Proteobacteria_unclassified, Alphaproteobacteria_unclassified, and Sphingomonadaceae_unclassified), Acidobacteria (genus Solibacteria), Actinobacteria (genera Streptomyces, Nocardia, Blasococcus, Corynebacteria, Solirubrobacteracea _unclassified, Micrococcaceae_unclassified, Actinobacteria_unclassified, Gaiella, and Pseudonocardiaceae_unclassified) and Cyanobacteria (genera Nostocales_unclassified, Leptolyngbyaceae_unclassified and Oxyphotobacteria) strongly associated with T1 as compared with control (Figure S5a, Table S5).
Fungal indicator species analysis at TL-stage identified OTUs from phyla Ascomycota (genera Acremonium, Aspergillus, Chaetomium, Colletotrichum, Fusarium, Helicoma Lecythophora, Microascus, Ochroconis, Penicillium, Preussia, Mycoleptodiscus, Ochroconis, Paecilomyces). Basidiomycota (genera Conocybe, Cryptococcus, Oliveonia, and Cryptococcus), Chytridiomycota (Kochiomyces, Rhizophlyctis, and Spizellomyces) strongly associated with T1 treatment. Conversely, members of the phyla Glomeromycota (genera Claroideoglomus, Paraglomus, Rhizophagus, Claroideoglomus, and Paraglomus) and Zygomycota (genera Mortierella and Rhizopus) were highly and exclusively associated with control samples (Figure S5b, Table S6).
At flowering stage, in T2 there was an increased representation of bacterial indicator species related to phyla Actinobacteria (genera Nocardioides, Actinobacteria_unclassified, Pseudonocardiaceae_unclassified, Conexibacter, Microtrichales_unclassified, Solirubrobacterales_unclassified, Janibacter, Pseudarthrobacter, Frankiales_unclassified, Microtrichales_ge), and Firmicutes (genera Planococcaceae_unclassified, Bacillales_unclassified, Oceanirhabdus, Clostridiaceae_1_unclassified, Tumebacillus, and Periconia). Cyanobacteria's OTUs were absent in control but present in T1 and T2. However, bacterial OTUs of phyla Proteobacteria (genera Rhizobiales_unclassified, Acetobacteraceae_unclassified, Myxococcale_unclassified, Caulobacteraceae_unclassified, Deltaproteobacteria_unclassified, and Nitrobacter) and Verrucomicrobia (genus Verrucomicrobia) showed a higher relative abundance in control compared to T1 and T2 (Figure 4a, Table S7).
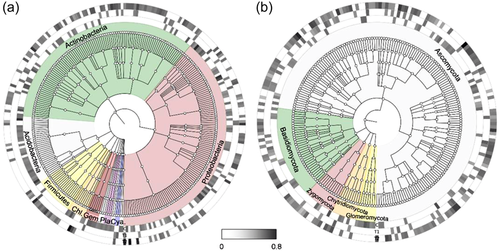
The fungal indicator community in T2 exhibited a higher relative abundance of OTUs related to Ascomycota (genera Exophiala, Claroideoglomus, Madurella, Funneliformis, Pyrenochaetopsis, Paecilomyces, Myceliophthora, Mortierella, and Gibberella etc.), Basidiomycota (genera Mortierella, Leptoxyphium, Penicillium, Aspergillus, and Coniochaeta) and Chytridiomycota (genus Acremonium). Interestingly, at TL-stage, members of Glomeromycota were absent in T1 and T2 but represented in control samples (Figure 4b, Table S8).
3.7 Structural equation modelling
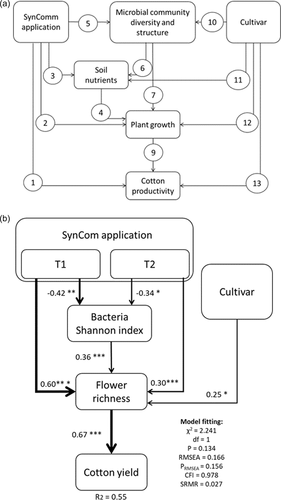
A rationale for the model is provided in the conceptual model (Figure 5a). SEM analysis showed that the SynCom application influenced cotton productivity (flower numbers). Among SynCom treatments T1 showed higher effects on the flowering and final yield as compared to T2, although both treatments positively induced higher flowering and yield. Plant flower number was impacted indirectly via changes in bacterial alpha diversity. SEM demonstrated that increase in flowering was positively linked with increase in final plant yield. The best SEM model also retained cultivar as the additional explanatory variables for cotton productivity, while eliminating all other soil nutrient and plant growth measures (plant height, root length, nitrogen, and phosphorus concentrations) (Figure 5b).
4 DISCUSSION
4.1 Effect of SynCom application on plant growth
This study showed that the seed application of SynComs improved germination and early plant growth parameters. Enhanced germination percentage and rate in T1 treatments (SynCom-seed dressing) suggests that SynCom application helped to break seed dormancy, possibly by quickening metabolic processes (Garcia-Lemos et al., 2020; Rostamikia et al., 2016). Likewise, higher shoot growth parameters (7.4% higher shoot height and 5.1% higher shoot biomass) indicated that SynCom application at seedling stage can confer improved plant fitness at the early growth stage. The functional traits of the SynCom members used in this study included IAA and NH4 production, and P-solubilisation, suggesting that these microbes were able to provide effective plant-growth-promoting phenotypes and hence increased plant performance. This finding is consistent with previous studies reporting that phytohormone production and higher acquisition of N and P could improve germination and early plant growth (Mukherjee et al., 2020; Pereg & McMillan, 2015; Trivedi et al., 2011) Increased performance of above-ground plant characteristics (plant height and biomass) at early growth stage supports our first hypothesis.
However, the early positive impacts of SynCom on plant height and root elongation were not apparent at the later stage of the development. It is possible that soil native microflora outcompeted the introduced SynCom (Pereg & McMillan, 2015). Alternatively, it is also possible that, while SynCom is still effective, plants growth parameter stabilised at later stage because plant invested heavily in reproductive structures (Berg et al., 2016; Paul et al., 2011; Pereg & McMillan, 2015) The second explanation is supported by increased flower numbers and seed yields in this study. Our results are consistent with earlier findings (Gomathy et al., 2008; Liu et al., 2022; Narula et al., 2005), which reported the impact of PGPR on flowers and yield.
Plant yield data showed that T1 and T2 elicited 10.4% and 7.4% higher yields than control plants. Interestingly, the early application of SynCom (T1) showed a higher yield than SynCom-soil treatment (T2), suggesting the role of priority effects on colonisation and host functions consequent to SynCom application. It further suggests that early plant fitness and microbial recruitment exert greater positive impacts on yield. Indeed, the SEM model demonstrated that alteration in bacterial alpha diversity and fungal beta diversity by the SynCom contributed to enhance flowering, which was positively associated with final cotton yield. Our findings are supported by recent studies that reported links between microbial community structure and plant productivity in different crops (Khan et al., 2019; Lebeis et al., 2015; Liu et al., 2022).
Most treatments showed significant effects on plant growth and productivity traits. However, in plant parameters such as plant height and root length, there were interactive effects of SynCom treatment cotton varieties. Particularly, at the true leaf stage, T1 promoted plant height in V1 and root length in V5 cultivar. In terms of flowering and plant productivity, V4 cultivar showed most positive response to SynCom treatment T1. This finding showed that PGPR as SynCom can also be adopted as potential a biological solution to achieve cultivar-specific gains in farming practices. Our findings are consistents with previous reports of interactions between PGPR consortia and crop genotypes to enhance specific plant growth and yield-related agronomic traits in crops for example chickpea and wheat (Imran et al., 2015; Pagnani et al., 2020; Tabassum et al., 2017).
4.2 Effect of SynCom application on rhizosphere microbes
Alpha diversity (Chao1 and Shannon index) of bacteria declined with SynCom application. This indicates that the SynCom application has likely modulated the colonisation patterns by native microflora over time. Our findings are inconsistent with previous findings that reported a positive effect of PGPR on microbial diversity (Zhang et al., 2019). This contradiction can be explained by the mode of application. Our seed dressing likely provided the introduced SynCom with a priority effect to colonise plant roots as soon as seed germinated (priority effect). Moreover, SynComs had been previously reported to help plants recruit beneficial microbes and thus indirectly promote growth (Nelson, 2018). SynCom seed dressing also affected bacterial community structure. Indicator species analysis showed that SynCom application was highly associated with higher recruitment of bacterial OTUs from Bacillales, Solibacterales, Nostocales, and Proteobacteria taxa. This suggests that SynCom PGPR are facilitating diverse groups of plant beneficial microbes to colonise plant roots. For example, a number of Proteobacterial taxa are known producers of plant hormones, N-fixation, and other PGPR activities (Nagel et al., 2018). Similarly, Bacillales and Actinobacteria members are known to provide pathogen protection as well as nutrient solubilisation (Suela, Silva et al., 2013). We also observed that our bacterial-based SynCom shifted fungal community structure in the rhizosphere. Particularly, SynCom seed dressing (T1) showed a strong effect at the TL-stage. It is possible that SynCom directly impacted fungal community. Alternatively, SynCom-induced shifts in bacterial communities could have indirectly elicited changes in the fungal community, as observed in previous research (Overbeek et al., 2021; Zhang et al., 2019). For example, enhanced recruitment of nonpathogenic fungal groups and Bacillus spp. abundance in the rhizosphere has been reported following PGP Streptomyces spp. inoculation (Yang et al., 2021). Further, the introduction of Bacillus velezensis NJAU-Z9 was reported to shift bacterial and fungal communities and increase yield (Zhang et al., 2019), which is consistent with our findings. Interestingly, we found that at TL-stage, a fungal OTU belonging to Fusarium sp. was significantly lower in SynCom-treated soils. A number of Fusarium species are well-known plant pathogens, suggesting that SynCom may provide biocontrol functions at early stage of plant growth, but this has yet to proven. Interestingly, the fungus Gibberella spp, which are known for production of gibberellins (Hedden & Sponsel, 2015), was found associated with T1 and T2 but was absent in the top 10 indicator OTUs of control samples. This supports our explanation that SynCom treatment can change fungal community structure to confer better plant fitness in some cases. This hypothesis is supported by our SEM analysis, which showed that a change in bacterial community with SynCom treatments was linked to higher number of flowers and thus directly associated with yield. This suggests that SynCom treatment has brought a change in plant performance and yield directly via providing nutrients and indirectly via changing rhizosphere microbial compositions and structure.
4.3 Effect of SynCom application on soil nutrient availability
Among the soil parameters, enhanced availability of soil nitrate under SymCom treatments at TL- and flowering stages indicates the possible involvement of Brevibacterium sp., a nitrogen fixing bacterium included in the SynCom tested in this study. It is also possible that SynCom encouraged recruitment of microbial groups that are involved in the nitrification and nitrogen fixation. Increased nitrate availability in T1 and T2 soils might also be linked to higher recruitment of Cyanobacteria, an important nitrogen fixing microbial group (Babu et al., 2015), which was strongly associated with both SynCom treatments in this study. Overall, the PGPR-induced increase in NPK availability in soils is well documented (Cordero et al., 2018). At the flowering stage, T2 was found more effective than T1 in terms of nitrate availability suggesting higher survival of nitrogen-fixing inoculants in soil drenched with SynCom treatments. However, the increased nutrient availability did not confer benefit in terms of plant performance in this case. Collectively, our findings suggest that early vigorous plant growth is more important for yield then later development or nutrient availability, and thus plants received most benefit from SynCom when it was applied as a seed dressing (Fukami, 2015; Zhang et al., 2019).
5 CONCLUSIONS
This study provides evidence for the positive impact of a SynCom on germination, plant shoot/root, and productivity. It also suggests that the seed dressing approach for SynCom application provides better outcomes than later stage soil treatment. The SynCom seed dressing can efficiently increase germination rate and early growth parameters. The cultivar and treatments both have differential growth responses and an impact on bacterial/fungal diversity. Notably, indicator species analysis showed different patterns, especially higher recruitment of Bacillales and Actinobaceriales by SynCom seed dressing. Overall, this study suggests that the use of SynCom can increase crop performance in cotton directly via provision of nutrients and hormones, and indirectly via manipulating microbial community structure in rhizosphere. If these findings can be replicated under field conditions, SynCom can provide an effective and complementary tool to conventional farming to increase farm productivity in an environmentally sustainable way.
AUTHOR CONTRIBUTIONS
Simranjit Kaur and Brajesh K. Singh designed the experiment. Simranjit Kaur performed the experiment and collected data and wrote the manuscript with significant inputs from all co-authors. Eleonora Egidi, Zhiguang Qiu, and Juntao Wang helped in microbial community analysis. Brajesh K. Singh, Catriona A. Macdonald, Eleonora Egidi, and Zhiguang Qiu provided guidance to undertake work. Pankaj Trivedi, Jay Prakash Verma, and Hongwei Liu provided with the microbial materials and preliminary data. All authors provided feedback on the manuscript.
ACKNOWLEDGEMENTS
Plant microbiome and microbial colonisation work in BKS lab is supported by the Australian Research Council (DP190103714; DP210102081). Postgraduate fellowship for SK was provided by Western Sydney University as a co-investment for the project funded by Cotton Research and Development Corporation. EE is supported by the Australian Research Council DECRA Fellowship (DE210101822).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
The authors confirm that they have followed the ethical policies of the journal.
Open Research
DATA AVAILABILITY STATEMENT
All data will be publicly available.




