Global and regional circulation trends of norovirus genotypes and recombinants, 1995–2019: A comprehensive review of sequences from public databases
Abstract
Human noroviruses are the leading global cause of viral gastroenteritis. Attempts at developing effective vaccines and treatments against norovirus disease have been stymied by the extreme genetic diversity and rapid geographic distribution of these viruses. The emergence and replacement of predominantly circulating norovirus genotypes has primarily been attributed to mutations on the VP1 capsid protein leading to genetic drift, and more recently to recombination events between the ORF1/ORF2 junction. However, large-scale research into the historical and geographic distribution of recombinant norovirus strains has been limited in the literature. We performed a comprehensive historical analysis on 30,810 human norovirus sequences submitted to public databases between the years 1995 and 2019. During this time, 37 capsid genotypes and 56 polymerase types were detected across 90 different countries, and 97 unique recombinant genomes were also identified. GII.4, both capsid and polymerase, was the predominately circulating type worldwide for the majority of this time span, save for a brief swell of GII.17 and GII.2 capsid genotypes and a near-total eclipse by GII.P16, GII.P21 and GII.P31 beginning in 2013. Interestingly, an analysis of 4067 recombinants found that 50.2% (N = 2039) of all recorded sequences belonged to three recently emerged recombinant strains: GII.2[P16], GII.4[P31], and GII.4[P16]. This analysis should provide an important historical foundation for future studies that evaluate the emergence and distribution of noroviruses, as well as the design of cross-protective vaccines.
Abbreviations
-
- HIV
-
- human immunodeficiency virus
-
- nt
-
- nucleotide
-
- ORF
-
- open reading frame
-
- VP
-
- virion protein
1 INTRODUCTION
Acute gastroenteritis is a major cause of morbidity and mortality worldwide, particularly in infants and young children from developing countries. Following the successful implementation of rotavirus vaccines, noroviruses have become the most important cause of viral gastroenteritis in the US and other developed countries.1 Norovirus disease is spread by contact with contaminated food or infected individuals, with intense symptoms of nausea, diarrhoea and abdominal pain that arise 12–48 h after initial infection. It is estimated that noroviruses annually cause up to 200,000 deaths and approximately $60 billion in financial burdens associated with health care and productivity loss worldwide.2, 3
One of the major obstacles on the development of an effective vaccine is the extensive genetic and antigenic diversity presented by noroviruses. Based on genetic differences on the major capsid protein (VP1), noroviruses have been classified in 10 genogroups (GI-GX) and multiple genotypes. Humans can be infected with multiple different genogroups (GI, GII, GIV, GVIII, GIX) and genotypes (>30).4, 5 While a single genotype, GII.4, has been predominant in humans for over 2 decades, other genotypes can cause large outbreaks and predominate in different locations. One example is the large number of outbreaks reported during 2014–2017 associated with GII.17 and GII.2 viruses.6-10 Because noroviruses present a hotspot of recombination at the boundary of the open reading frames coding for the non-structural (ORF1) and capsid (ORF2) proteins, a dual typing system is used to describe circulating viruses.4 Thus, ORF1/ORF2 recombinant viruses are designated with numbers representing their capsid and polymerase types (e.g. GII.2[P16], GII.6[P7]).
The emergence of different predominant noroviruses has been associated with changes on VP1, which allows for viruses to escape immunity developed against previous infections.5 However, this notion has changed in recent years as most predominant noroviruses are all linked to changes on the non-structural proteins and recombination events (e.g. GII.4[P31], GII.2[P16]). Notably, the emergence of a novel GII.P16 ORF1 allele has been linked to multiple new recombinant strains circulating worldwide (e.g. GII.4[P16], GII.2[P16]; GII.12[P16]).5, 9-13 Although theoretically over 1200 different capsid-polymerase type combinations are possible, large genome studies showed that only a fraction of those combinations seem to be viable and persist in nature.14 While there is extensive literature dedicated to describing the importance of recombinant norovirus in human gastroenteritis, there are limited studies that quantitatively evaluate the global distribution of recombinant strains over long time periods.
In this study, we harnessed all sequences available in public repositories of noroviruses circulating from 1995 to 2019 to provide a comprehensive analysis of human norovirus genotype circulation and predominance at global and regional level. We also provide a historical perspective on how norovirus genome sequencing has evolved and, just recently, improved. We hope these data will provide a baseline for studies that evaluate the distribution of viruses and the design of cross-protective vaccines.
2 METHODS
2.1 Norovirus dataset curation and cleaning workflow
A total of 45,095 norovirus sequences exceeding the length of 100 nucleotides (nt) and available between the years 1995 and 2020 were downloaded from GenBank (https://www.ncbi.nlm.nih.gov/GenBank/) on 2 February 2021. The sequences were downloaded in FASTA format and were evaluated for GI or GII polymerase and capsid genotype identity and sequence coverage using the Norovirus Typing Tool (https://www.rivm.nl/mpf/typingtool/norovirus/).15 Genotype identity, sequence coverage, and other relevant metadata were retained and organised in RStudio. Virus sequences lacking sufficient sequence coverage to type either region of interest were discarded from the data set. The remaining virus sequence data (N = 43,999) were paired with relevant metadata (strain, organism, isolate, year of isolation, country, host, isolation source, publication title) available in the GenBank record. In instances where year and country of isolation metadata were not listed in one of the GenBank tags, such information was manually parsed from the strain or isolate tags where available. Viruses with ambiguous or missing year or location metadata were discarded from the dataset. Noroviruses collected during 2020 were omitted to only encompass 25 years of study (1995 and 2019).
To sort for human noroviruses, the data set was filtered for all instances of host metadata associated with Homo sapiens (‘Homo sapiens’, ‘Human’, etc). In instances where host metadata was not independently listed in GenBank tags, isolation source data was manually evaluated with the same key words. Viruses with missing host metadata were discarded from the dataset. Additionally, this dataset of human noroviruses was also screened for indications of immunocompromised hosts in the host, isolation source and title metadata (e.g., ‘Immunocompromised’, ‘HIV positive’), which were omitted upon detection. The final dataset totalled 30,810 human noroviruses between the years 1995–2019. A flowchart of the data cleaning process can be found in Figure 1a.
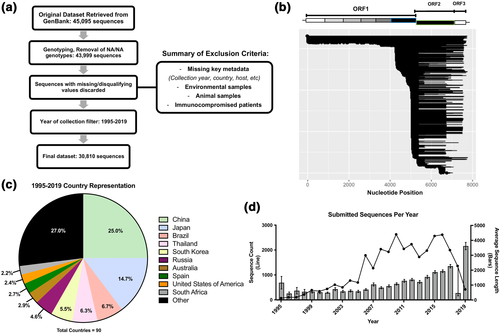
Development and evaluation of a comprehensive global norovirus dataset. (a) Flowchart process of the initial procurement of 45,095 norovirus sequences collected between the years of 1995–2020, followed by independent genotyping of the capsid and polymerase regions via the online genotyping tool. Sequences that were unable to be genotyped were discarded, and stringent criteria were used on the remainder to filter the dataset to only contain human infecting noroviruses with verified collection metadata (year and country of isolation, etc). Following the exclusion of disqualified sequences and a year filter of the dataset to only encompass the year range of 1995–2019, a total of 30,810 human norovirus sequences remained for further analysis. (b) Visualization of the sequences in the final dataset aligned to a schematic of the norovirus genome. The genome regions associated with genotyping of the RNA-dependent RNA polymerase and capsid are outlined in blue and green respectively. (c) Breakdown of human norovirus sequence submissions between the years 1995-2019 by country. The respective contributions by the 90 recorded countries are presented by the top 10 most represented countries, with the remaining 80 condensed as Other. (d) Assessment of data richness of norovirus sequences submitted to GenBank by year. The dotted line (left y-axis) denotes the total sequences submitted to GenBank each year. The bars (right y-axis) show the average sequence length of submitted sequences each year, with the bars denoting a 95% confidence interval
2.2 Data visualization
Statistical analysis and data visualization for the large-scale datasets were conducted in RStudio using tidyverse and associated packages, as well as Prism7 software. Geographic distribution of the human noroviruses was visualised in R using the maptools package. Administrative map shape file (1:10 m Cultural Vector, Admin 0 – Countries) was obtained from Natural Earth website (https://www.naturalearthdata.com/downloads/10m-cultural-vectors/10m-admin-0-countries/, accessed on 25 June 2021), and country boundaries were dissolved into eight geographic regions: North America, Latin America and Caribbean, North Africa and Middle East, Sub-Saharan Africa, Europe and Central Asia, East Asia and Pacific, South Asia, and Oceania. The number of sequences of each genotype in each region was counted and summarised in the pie charts using mapplots package. Heatmap order of polymerase types and capsid genotypes was derived from sequence alignments using MEGAX software from FASTA files containing up to two random representative sequences for each respective genogroup.14, 16, 17
3 RESULTS
3.1 A quantitative and qualitative assessment of archival norovirus data
After stringent filters across multiple metadata parameters were applied to the original sequence database (N = 45,095), we obtained a final dataset comprised of 30,810 human norovirus sequences collected between the years 1995 and 2019 (Figure 1a). According to GenBank records, 16,972 (55.1%) of these sequences were reported in publications listed in PubMed. The length of these sequences spanned a range between 101 and 7778 nt, with a mean length of 961 nt and a median of 293 nt. On average, sequencing coverage begins around nucleotide position 4683 on the genome, allowing for typing of either the polymerase region, the capsid region, or both. The distribution of sequence coverage in this dataset with respect to the norovirus genome can be found in Figure 1b.
The spatiotemporal distribution of the dataset was also evaluated to better contextualise our investigation. A breakdown of archival norovirus distribution by country can be observed in Figure 1c, organised as the top 10 most represented countries with the remainder grouped in the Other category. While 90 countries were represented by this dataset, the distribution was highly skewed, with over 51% of all virus sequences contributed by the top four countries (China = 7690, Japan = 4551, Brazil = 2059, Thailand = 1948). Moreover, countries on the lower end of the distribution were largely underrepresented in terms of norovirus collection data, with 18 countries attributed to 10 or fewer norovirus sequences deposited into GenBank over a 25-year period. As a consequence, despite the necessity and utility of describing annual circulation trends at a global level, these results will be inherently biased from the overrepresentation of certain countries. Thus, it was additionally prudent to bin subsequent geographic distribution analyses into discrete regions.
The breadth and depth of norovirus sequences collected between the years 1995–2019 were also examined (Figure 1d). It was observed that only a minimal number of norovirus sequences were collected between the years 1995 through 1999. While modest increases in contribution to the norovirus record were observed in the years following 2000, it was not until 2006 that over 1000 sequences were recorded in GenBank in a single year. These annual trends have remained largely consistent, peaking in 2016 with 2615 reported norovirus sequences. Additionally, a steady increase in the length of sequence coverage has been observed over time, with the average length of submitted sequences consistently covering 1000 nt or more since 2011.
Lastly, given the broad variety of sources collected for this study, it was prudent to verify whether any singular source or outbreak contributed a disproportionate number of submitted sequences to the dataset. An analysis of sequence contribution from 1124 unique titles showed that 98.0% of these sources contributed less than 1.0% to the dataset, with 77.6% of sources accounting for ≤0.01% (Figure S1A). Only nine sources were identified to individually exceed 1.0% sequence contribution, with a single outlier accounting for 3.93% of total sequences. However, a closer examination of per-year sequence contribution from these sources showed that they were the output of multi-year surveillance programs, spanning between two to 14 years in scope (Figure S1B). As such, it is unlikely that these identified sources introduce a concerning degree of skew to the total dataset.
3.2 Spatiotemporal distribution of archival norovirus polymerase and capsid genotype circulation
A total of 25,632 norovirus sequences from the sampled dataset presented sufficient coverage for successful genotyping of the capsid region. Thirty-seven unique capsid genotypes were detected from the curated dataset, with the top 10 most prevalent displayed in Figure 2a. Interestingly, the top 10 list of most prevalent annually circulating genotypes contained at least one representative genotype from nearly all norovirus immunotypes.18 GII.4 was the predominant circulating capsid genotype, accounting for 52.6% of all available sequences between the years of 1995–2019. The second most prevalent genotype was GII.3 at 11.5%, followed by GII.2, GII.17, GII.6, GI.3, GII.13, GI.4, GII.12, and GII.14. The remaining capsid genotypes accounted for only 8.4% of the total sequences. An analysis of the annual circulation of capsid genotypes shows a consistent presence of GII.4, accounting for close to 60% of all recorded sequences at certain years (Figure 2b). GII.3 was also present across all measured years at a consistent frequency (range of 3.9%–25.0%, with an average of 11.7%). In contrast, while circulation of GII.2 and GII.17 was observed at low levels in earlier years (respective ranges of 1.0%–7.5% and 0.0%–6.0%), there was a marked increase in prevalence of both genotypes in the last 6 years, with GII.2 becoming the predominant circulating genotype in 2017, accounting for 47.9% of all recorded sequences that year. To better visualise these granular changes, a second graph of annually circulating genotypes was constructed with GII.4 data omitted (Figure S2).
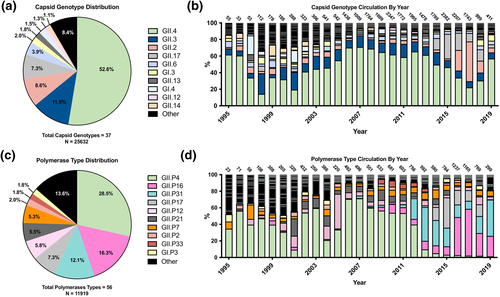
Temporal distribution of circulating norovirus capsid genotypes and polymerase types between the years 1995-2019. (a) Genotype identification and quantification of the 25,632 norovirus sequences that sufficiently covered the capsid region. The top 10 represented genotypes are displayed, with the remainder consolidated into an Other category. (b) Percentage breakdown of globally circulating capsid genotypes by year. Colour coding of capsid genotypes are the same as those established in Figure 2a, though the Other category has been subdivided to show the number of genotypes comprised within a given year. The number of sequences for each year are displayed at the top of each respective bar. (c) Type identification and quantification of the 11,919 norovirus sequences that sufficiently covered the polymerase region. The top 10 represented types are displayed, with the remainder consolidated into Other. (d) Percentage breakdown of globally circulating polymerase types by year. Colour coding of polymerase types are the same as those established in Figure 2c, with Other category subdivided to show the number of polymerases comprised within a given year. The number of sequences for each year are displayed at the top of each respective bar
Given the enduring predominance of GII.4 and its pronounced epochal diversification,5, 19-23 we further analysed the temporal distribution of pandemic variants (Figure S3). An analysis of 13,483 GII.4 sequences between the years of 1995–2019 recapitulated the chronological emergence and replacement of variants previously reported in the literature,5, 22, 24 with the current variant Sydney_2012 maintaining predominance for a longer span of years than any of its previous counterparts. It is worth noting that a small contingent of GII.4 sequences were either unable to be assigned a variant or were assigned one outside the boundaries of commonly accepted circulation years. This was mostly observed for short (100–300 nt) sequences from highly conserved regions of the capsid protein that provided insufficient phylogenetic signal for variant identification.
A total of 11,919 norovirus sequences from the sampled dataset presented sufficient sequence coverage for polymerase typing. Quantification of the 56 unique polymerase types also saw GII.P4 as the predominant circulating type, albeit at only 28.5% of total recorded sequences (Figure 2c). GII.P16 and GII.P31 (formerly, GII.Pe) polymerases were the next most frequent at 16.3% and 12.1%, respectively. The rest of the top 10 most frequent types was comprised of GII.P17, GII.P12, GII.P21 (formerly, GII.Pb), GII.P7, GII.P2, GII.P33 (formerly, GII.Pg), and GI.P3, with all remaining types accounting for only 13.6% of available sequences. Interestingly, the temporal distribution of polymerase type circulation showed that while GII.P4 initially circulated at similar levels to its capsid counterpart, a steady decrease in prevalence beginning in 2007 gave way to a near-total replacement in 2013 by GII.P16, GII.P31, and GII.P17, which have alternated as the predominant polymerases between the years 2013–2019 (Figure 2d).
Following the observation that some countries have contributed a vastly disproportionate quantity of archival norovirus sequences compared to others, it was necessary to organise the global circulation of genotypes into eight distinct geographic regions: North America, Latin America and Caribbean, North Africa and Middle East, Sub-Saharan Africa, Europe and Central Asia, East Asia and Pacific, South Asia, and Oceania (Figure 3). While sample sizes varied greatly between regions, the cumulative analysis of circulating capsid genotypes largely reflected the previously described global trends. GII.4 was the predominant capsid genotype in all regions, accounting for half or more of all recorded sequences in each area with a minor exception in South Asia. Fluctuations in frequencies between the other top 10 capsid genotypes were also observed at a regional level, such as the higher prevalence of GII.2 over GII.3 in North America. Additionally, a larger presence of the GII.6 genotype was also seen in the Latin America and Caribbean region, as well as in Europe and Central Asia.
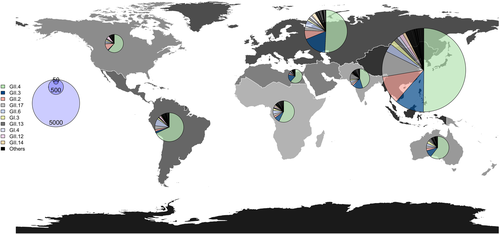
Regional distribution of circulating capsid genotypes between the years 1995-2019. World map subdivided into eight geographic regions: North America, Latin America and Caribbean, North Africa and Middle East, Sub-Saharan Africa, Europe and Central Asia, East Asia and Pacific, South Asia, and Oceania. The distribution of the top 10 circulating capsid genotypes from Figure 2a are displayed for each region. The sizes of the pie charts denote the number of norovirus sequences sampled for each respective region
Additional regional differences were detected when we visualised the temporal distribution of the respective changing frequencies of circulating capsid and polymerase types (Figure S4 A, B). For example, the increased presence of GII.2 between the years 2015–2019 was mostly from North America, Europe and Central Asia, and East Asia and Pacific regional circulation trends. Additionally, some circulation patterns were entirely unique to particular regions, such as the overwhelming majority of GII.13 circulation occurring in South Asia. Regarding polymerase type circulation, all regions presented the shift in predominance from GII.P4 to that of GII.P16, GII.P31, and GII.P17 that began in 2010–2014. However, the respective proportions of these genotypes varied greatly between regions. GII.P16 circulation was predominant in the North America, Latin America and Caribbean, Europe and Central Asia and South Asia regions, while GII.P31 eventually became the predominant polymerase genotype circulating in the North Africa and Middle East and Sub-Saharan Africa regions. In contrast, GII.P17 circulated at high levels in the East Asia and Pacific region as well as Europe and Central Asia and was the predominant genotype of Oceania during the years 2015–2019. Some of these data trends could be attributed to the extremely limited sequence collection of certain regions, particularly during the years prior to 2007, which complicates the interpretation of these results.
3.3 Incidence and frequency of genotype recombination events
Recent studies have indicated that recombination events between polymerase and capsid regions may be a significant driver in the emergence and predominance of norovirus genotypes and variants.25-27 Thus, we analysed the dataset for the incidence of archival recombinants to provide a quantitative description of the role of recombinant strains on human norovirus diversity. Only 6471 (21.9%) sequences were able to be genotyped for both the polymerase and capsid regions; 4067 of these (62.8%) sequences were determined to be recombinants. The temporal distribution of this dataset showed many years with under 100 double-typed genomes until 2007, complicating analysis (Figure 4a). However, the proportion of recombinant sequences also rose dramatically alongside these increased sampling trends, with recombinant strains accounting for the majority of double-typed sequences from 2012 onward.
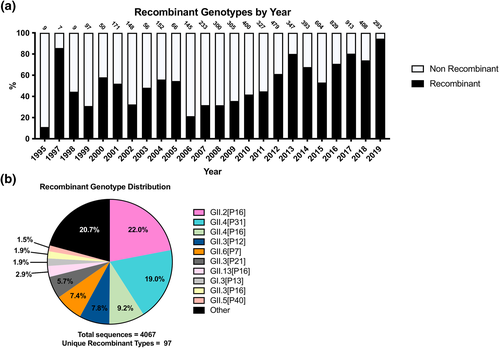
Incidence and frequency of circulating recombinant norovirus strains. (a) Ratio of recombinant (black) to non-recombinant (white) noroviruses by year for the 6471 norovirus sequences with sufficient sequence coverage. The number of double-type sequences for each year are listed in the numbers above each respective bar. (b) Double-typed frequency of the 4067 recombinant norovirus sequences. The top 10 most frequent polymerase/capsid types are featured in this pie chart, with the remaining 87 instances of unique recombination events consolidated into Other
While 97 unique recombinant strains were uncovered from the analysis, three account for half (∼50.2%) of the 4067 total sequences: GII.2[P16], GII.4[P31] and GII.4[P16] (Figure 4b). Notably, these recombinants correlate with the recent emergence and rapid proliferation of capsid genotype GII.2 and polymerase types GII.P16 and GII.P31. Indeed, recombination events with GII.P16 account for four of the top 10 most represented recombinant genomes. In contrast, the remaining 87 recombinant strains only represented 20.7% of all analysed sequences, with many of these only accounting for a single recorded instance of recombination. Beyond the predominant recombinant strains, another point of interest was elucidating how recombination events might aid in perpetuating the long-term circulation of less prevalent polymerase types. For example, the circulation of polymerase type GII.P12 is characterised by a large spike in prevalence between the years 2004–2005, followed by moderate levels of circulation to present day (Figure S5A). To the degree that double-typed sequences are available for this time, these trends are associated with recombination events with GII.4 and GII.3, respectively (Figure S5C). In contrast, GII.P21 circulation is characterised by multiple boom-and-bust cycles between the years 2000–2019, which also seems to be associated with the relative levels of the GII.3[P12] recombinant strain (Figure S5B, D). Taken together, these data suggest that recombination events may not only be responsible for the sudden appearance of major shifts in genotype predominance but may also play a role in the prolonged persistence of minor genotypes circulating in the background.
Potential limitations to recombination types have been recently described using nearly-full norovirus genome sequences.14 Accordingly, we assembled a heatmap for all double-typed sequences in our dataset to better assess the full scope of all norovirus recombination data available on GenBank (Figure 5). Broad analysis showed a far higher incidence of recombination events within GII than GI. Despite a single report from a study in India,28 there was no single incidence of recombination events between GI and GII genogroups. Unexpectedly, two instances of recombination events between GII and GVIII or GIX viruses were also identified. However, the parental viruses for GVIII and GIX have not been fully described, so the proper nature of these discrepancies on the phylogenetic clustering remain uncertain. Many of the polymerases identified in the breakdown of the most prevalent recombinant strains were shown to associate with five to 10 different capsids. In particular, the GII.P16 polymerase associated with 10 different capsids (including its nonrecombinant GII.16 counterpart), though recombination events with GII.2, GII.4 and GII.13 made up the majority of recorded double-type sequences. In contrast, polymerases such as GII.P31 had recorded associations with five capsid genotypes, but the recombinants with GII.4 accounted for the overwhelming majority of all double-typed sequences. Shifting to capsid genotypes with broad polymerase associations, our analysis found that eight capsids had documented associations with six or more polymerase types. Of these, GII.2 and GII.3 were associated with 10 and eight different polymerase types (including non-recombinants), with the most prevalent recombinant pairings being GII.2[P16] and GII.3[P21], respectively. A breakdown of the top 10 capsid and polymerase types with the highest number of associations with their counterpart regions can be found in Figure 5b, c.
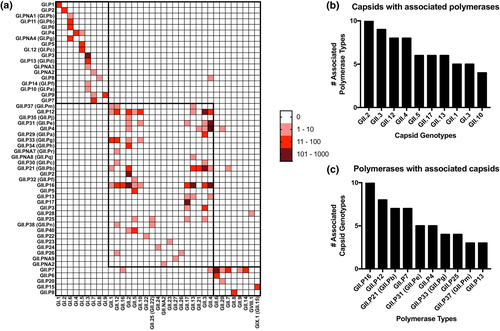
Heat map of norovirus polymerase and capsid recombination events. (a) Heat map comprised of all double-typed norovirus sequences, organised by polymerase genotype (y-axis) and capsid genotype (x-axis). Red-colored squares indicate the incidence of a double-typed sequence, while the intensity of the colour denotes the quantity in which a given sequence is found in the dataset (0, 1–10, 11–100, 101–1001). The bold boxes on the heatmap partition the data into Genogroups I and II, and further divide the latter into phylogenetic subgroups defined in Tohma et al. (2021)14. (b) Bar graph of the top 10 capsid genotypes with the highest number of unique polymerase type associations. (c) Bar graph of the top 10 polymerase types with the highest number of unique capsid genotype associations
4 DISCUSSION
Norovirus is a major cause of acute gastroenteritis worldwide and major efforts have been undertaken for the development of vaccines and specific therapeutics to treat the disease. A major roadblock to the development of measures against norovirus disease is the extreme genetic diversity, which is generated by genetic point mutations and recombination events. In addition to the genetic variability, estimates of norovirus burden on the human population have been complicated by multiple factors, including its complex infection dynamics (e.g., major presence in asymptomatic individuals and prolonged presence after the symptomatic phase) and its major role in endemic and epidemic cases. All these factors hinder the assessment of total number of cases associated with gastroenteritis at the global level, as endemic cases are counted as individual cases while outbreaks are mostly associated with the diagnostics of a small sampling of cases for each event. Despite this, major efforts have been undertaken to generate networks that use standard sampling and diagnostic test to evaluate frequency, distribution, and genetic evolution of noroviruses.22, 29, 30 The largest endeavour was conducted by a group of laboratories under the umbrella of Noronet,22 which included the characterisation of >16,000 norovirus-positive samples from 19 countries to determine genotype dynamics at the global level from 2005 to 2016. In this work we consolidated 25 years of archival human norovirus data publicly available on genetic databases. The sequences and associated metadata of over 30,000 norovirus entries were exhaustively analysed to provide a portrait of the chronological emergence, persistence, and replacement of circulating genotypes and recombinant noroviruses at a global and regional scale.
A comprehensive survey of circulating norovirus genotypes since 1995 has shown the persisting predominance of the GII.4 norovirus capsid, which has accounted for close to half of all annually submitted capsid sequences worldwide. Notable exceptions to this trend have arisen since 2015, such as the rapid proliferation of the GII.17 and GII.2 genotypes (Figure 2a). An even more striking shift in predominance was seen among the polymerase types, in which GII.P4 predominance was replaced entirely by GII.P31, GII.P16 and GII.17 in the past few years (Figure 2b). To the degree that they are available, analysis of double-typed sequences indicate that these trends may be linked, and that recombination events have propelled the global distribution of these hitherto minor genotypes circulating in the background (Figure 5b). Additionally, recombination events may also account for more subtle trends in the norovirus record. For example, boom-and-bust circulations of the GII.P12 and GII.P21 (formerly GII.Pb) polymerase types correlate with the enduring circulation of the counterpart GII.3 genotypes in the recombinant strains (Figure S5). The reason for the emergence of recombinant viruses is still unclear, but it is likely to be a multifactorial event that facilitates viral replication, transmission, and immune escape.5
An important aspect of large-scale bioinformatics work is to describe and evaluate the state of the existing archival record. To that end, we note the vast increase of norovirus sequences annually deposited into GenBank, as well as the improved fidelity of sequence coverage, which began around 2006 and has continued to present day (Figure 1d). Many factors potentially contribute to this rise, from a shift in academic interest to the field of noroviruses,1 to the increased cost effectiveness of high-fidelity sequencing, to the global accessibility of online digital archives. Conversely, all years prior to 2006 are marked by diminishing contributions to the archival record, with fewer than 100 sequences deposited during the earliest cut off year of 1995. While the initial download of this dataset contained even older norovirus sequences dating back to the 1960s, it was determined that the record for these years was too small and incomplete to draw any meaningful conclusions regarding genotype circulation. Indeed, even with the 1995–2019 timeframe, the norovirus record from many geographic regions is severely underrepresented or even non-existent until at least the year 2000 (Figure S4). Lastly, at the time of sequence download, a sharp decline in submitted norovirus sequences was observed after 2017, with only 437 sequences deposited in 2019. However, it is unclear whether this denotes a legitimate downturn in submitted sequences or of behavioural changes in the way norovirus sequences are reported in the literature and genomic databases.
A study of this nature has several inherent limitations beyond the technical aspects. While we were able to draw from a considerable number of independent resources for this study, there remains a large quantity of archival human norovirus sequences that have not been made available on public databases. Inferences about the circulation of norovirus genotypes and recombinants are further complicated by the infection dynamics of the virus itself. The existence of factors like global travel and asymptomatic individuals may introduce difficulties in the efforts to cohesively map the spatiotemporal distribution of circulating genotypes. Moreover, the differing diagnostic heuristics typically employed for endemic cases versus outbreaks, as well as surveillance programs for high-risk groups such as children and the elderly, may make raw sequence counts an unreliable metric for assessing shifts in norovirus predominance, and the integrity of additional collection metadata may be required for proper evaluation. Furthermore, the recent recognition of the importance recombination events play in the emergence and persistence of certain genotypes may paradoxically introduce a new source of bias into the norovirus record. That is, the enduring predominance of recombinant strains such as GII.4[P16] has sparked a concerted global research effort into that phenomenon that far exceeds any endeavours from prior years. Lastly, it is worth noting that when noroviruses were initially typed the unknown capsid and polymerase are assigned the same number (e.g. GI.1[P1], GI.2[P2]), and thus the lack of recombinant sequences in the early years of the record may be an artefact of the nomenclature. Since we do not have the whole evolutionary picture, the definition of recombinants is arbitrary to the nature of timing of identification and characterisation of sequences. Best examples of this phenomenon are shown by GII.2[P32], GII.4[P39], GII.5[P22], and GII.7[P36], which were first collected in the 1970s but retrospectively characterised in a recent study.14 Taken together, these concerns highlight the importance of retrospective analyses of archival samples wherever possible, as well as the need for partnerships between different laboratories and networks to converge global databases that would facilitate studies of norovirus transmission and diversification at the global level, similar to the efforts of the GISAID initiative.31 We hope this study will serve as a starting point for discussion related to the need for better databases that can be used to monitor and estimate the burden of norovirus to the human population, as well as aid in the development of cross-protective norovirus vaccines.
ACKNOWLEDGEMENTS
We would like to thank Dr. Lauren Ford-Siltz for critical reading and suggestions on this project. Financial support for this work was provided by the Food and Drug Administration intramural funds Program Z01 BK 04012 LHV (to G.I.P.).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
J.A.K was responsible for the acquisition of sequences from public databases, data curation, development of scripts for data parsing and visualization, critical analysis of the data and drafting of the original manuscript. K.T was responsible for data curation, development of scripts for geographic representation of genotypes, critical analysis of the data, reviewing and editing the manuscript. G.I.P. was responsible for conceptualization, funding acquisition, data curation, critical analysis of the data, reviewing and editing the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
A summary dataset of the 30,810 human noroviruses accession numbers, Pubmed IDs, country and year of collection, sequence length, and capsid and polymerase types has been made available with this study. A full dataset with all associated metadata can be obtained from the authors upon request.