Opposite allosteric mechanisms in TetR and CAP
Abstract
Regulation of the DNA binding affinity of an oligomeric protein can be considered to consist of an intrinsic component, in which the affinity of an individual DNA-binding domain is modulated in response to effector binding, and an extrinsic component, in which the relative position of the protein's two DNA-binding domains are altered so that they can or cannot contact both half-site operators simultaneously. We demonstrated directly that the TetR repressor utilizes an extrinsic mechanism and CAP, the catabolite activator protein, utilizes an intrinsic mechanism.
Introduction
Many of the DNA binding proteins that are involved in gene regulation are allosterically regulated in that their affinity for DNA is controlled by their binding of small molecules. Although allosteric proteins have been analyzed for more than 90 years,1-6 the mechanical details underlying allostery are still poorly understood in most systems.
An oligomeric DNA-binding protein could modulate its DNA binding affinity in two ways: intrinsically, by varying the intrinsic DNA binding affinity of the individual DNA binding domains, DBDs, and extrinsically, by varying the relative position or orientation of the two DBDs, so that both are or are not able to contact their DNA binding sites simultaneously (see Fig. 1). Although a protein could utilize both the intrinsic and extrinsic mechanisms to alter its DNA binding affinity, it seems likely that many proteins will be found to use primarily one mechanism or the other. It would at first appear that analysis of the relevant structures of a protein could reveal its mechanism for allosteric regulation of its DNA binding affinity. For example, in the case of a repressor that dissociates from DNA when effector is bound, one might expect to find that the structure of the repressor free in solution is very similar to the DNA-bound form, and that the effector-bound form differs from these. Even when the structures of the three requisite states are known, it is not yet possible to definitively determine the relevant allosteric mechanism just from the structures. The problem with a structure-based analysis is the difficulty in computing to the requisite accuracy the free energy differences between different structures of a protein in solvent. When a relevant structure must be inferred, the reliability of a structure-based deduction as to allosteric mechanism is even less secure. For the same reason, identification in biochemical experiments of structural differences between two states of a protein cannot definitively identify the protein's allosteric mechanism.
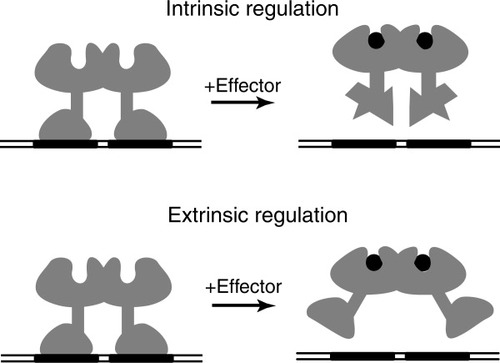
Mechanisms of allosteric regulation of a homodimeric DNA-binding protein. The drawing depicts a protein that is released from DNA by the binding of effector. Intrinsic mechanism: the intrinsic affinity of a DBD for its half-site operator is controlled by effector. Extrinsic mechanism: the separation or relative orientation of the two DBDs is controlled by effector.
In this work, we directly determined allosteric mechanisms utilized by two DNA-binding proteins whose affinity for DNA is dramatically affected by small molecule effectors. It is important that this information be available for future computational studies and analyses of the allosteric mechanisms of these proteins. The first protein was TetR, a repressor of the tetracycline resistance genes in Escherichia coli.7 The affinity of TetR for its operator DNA, tetO, is reduced significantly in the presence of its effector, tetracycline. The second protein was CAP, a transcription regulator of many genes in prokaryotes.8-10 Its affinity for its DNA binding site is increased in the presence of its effector, cyclic AMP. Each protein has been extensively characterized with genetic, biochemical, and biophysical experiments, but for each protein, the existing data can be interpreted as supporting either of the two possible allosteric mechanisms, see discussion. Here, we report experimental measurements on these proteins that provide direct evidence that TetR utilizes an extrinsic mechanism and that CAP utilizes an intrinsic mechanism.
Results
Principles for mechanism determination
We used two methods to determine allosteric mechanisms utilized by these proteins. The first, termed the half-site method, measures the affinity of a protein for its operator half-site. An intrinsic mechanism is involved if the affinity of a protein for a half-site operator, contacted by a single DNA binding domain, is changed by the presence of the protein's small molecule effector. If the affinity is unchanged by the effector, the mechanism must be exclusively extrinsic. Another way to identify the mechanism is to examine binding to special target DNA. In this DNA, the two DNA half-sites are flexibly connected in a way that allows the half-sites to assume any position and orientation required for unstressed binding of both DNA binding domains in the plus and the minus effector-bound states of the protein. If the protein's mechanism is exclusively extrinsic, that is, the DBDs move in response to effector, then the affinity of the protein for the linked half-sites will be independent of the presence of the effector, and if it is intrinsic, the affinity for the linked half-site DNA will vary with effector by the roughly the same amount that the affinity for normal DNA binding site varies, (see Fig. 2).
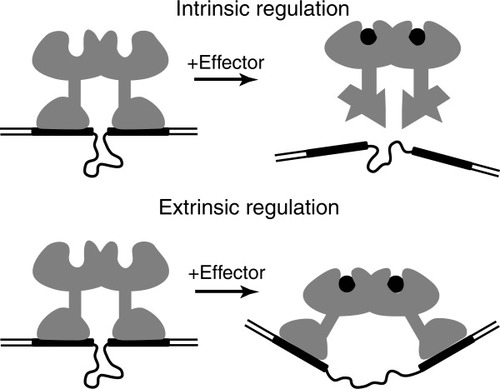
Method for determination of allosteric mechanism using the flexibly linked half-sites. If the mechanism is intrinsic, affinity for binding to the flexibly linked half-sites will respond to the small molecule effector in nearly the same way affinity for binding to normal operator. If the mechanism is extrinsic, the binding affinity will be nearly independent of effector.
Allosteric mechanism used by TetR
The half-site method for mechanism determination is straightforward, but its application requires the use of sufficiently high protein concentrations that binding to the half-site can be detected. Our attempts to use the half-site method with TetR were prevented by the relatively weak interaction between TetR and a half-site operator, with no binding observed at concentrations up to 2 μM TetR either in the presence or absence of tetracycline, and also by aggregation of the protein at high concentrations (data not shown). Therefore, for this protein we applied the linked half-site method.
Single-stranded DNA was used as the connector between the two double-stranded tetO half-sites. Biochemical and genetic evidence shows that tetO consists of two half-sites separated by a single non-contacted base pair.11, 12 Therefore, our single-stranded DNA linker needs to allow the two half-sites to approach as closely as ∼3–4 Å and must allow them to separate by at least 6–7 Å.13 We removed the central base on one strand, leaving a gap, and on the other strand, replaced the central base with one, three, or five thymine residues.
As assayed by the electrophoretic mobility shift assay of fluorescent-labeled DNA, the TetR used in these experiments possessed its normal behavior. Tetracycline binding to TetR dramatically reduced TetR's affinity for normal operator DNA, (see Fig. 3). Because the spacing between the two half-sites of T1-linked DNA is not likely to be significantly different from normal operator DNA, it is not surprising that for T1-linked half-sites, tetracycline binding to TetR substantially reduced TetR's binding affinity, (Fig. 4A).
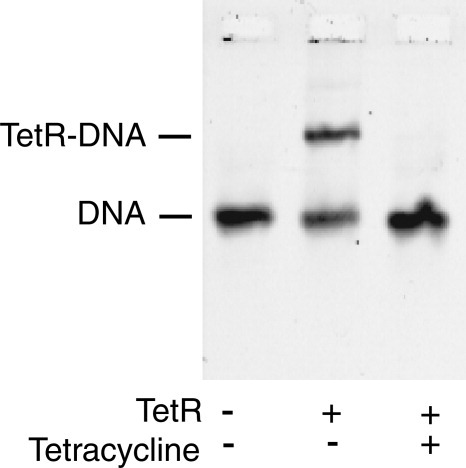
Tetracycline reduces TetR's ability to bind to full-site tetO.
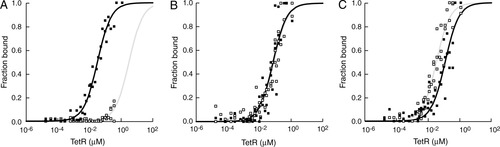
TetR binding to T1-linked (A), T3-linked (B), and T5-linked (C) tetO half-sites in the presence (open squares) and absence of tetracycline (black squares). The plots show the normalized fraction of DNA bound, and the fits to the Langmuir binding isotherm (solid lines).
Binding to the DNAs containing longer linkers was dramatically different from binding to normal operator or T1-linked half-site DNA. In 200 mM NaCl buffer and at binding equilibrium, TetR bound to the T3-linked DNA with nearly the same affinity (Fig. 4B) and to the T5-linked DNA with a slightly higher affinity (Fig. 4C) in the presence of tetracycline compared with its absence. If TetR's mechanism of allosteric regulation were exclusively intrinsic, then, regardless of the DNA used, for normal operator DNA or our T1-, T3-, or T5-linked DNAs, tetracycline would have dramatically decreased the DNA binding affinity. Instead, for the T3- and T5-linked DNAs, tetracycline produced almost no change. Thus, the mechanism used by TetR cannot contain a substantial intrinsic component and therefore, TetR must use an extrinsic mechanism. These results do not exclude, however, the existence of a minor intrinsic component in the allosteric response of TetR.
Allosteric mechanism utilized by CAP
We first demonstrated that the CAP protein used in this study was fully active. A 210-bp fragment of DNA containing a CAP binding site was amplified from pBAD-GFP plasmid and purified. An electrophoretic gel mobility shift assay was used to measure binding between this DNA and CAP, (see Fig. 5). As expected, CAP fully titrates the DNA in a 1:1 manner showing the CAP is essentially fully active. The binding affinity is too tight to measure the dissociation constant under these conditions. No binding was observed in the absence of cAMP (data not shown).
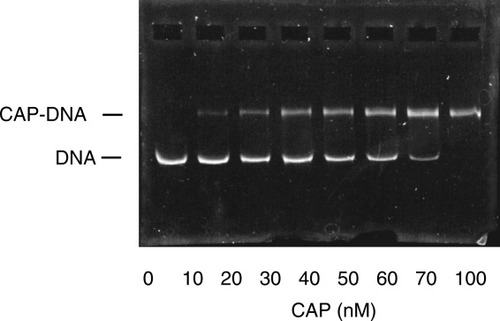
CAP binding to full-site DNA in the presence of cAMP.
We were unable to detect sequence-specific binding by CAP in the absence of cAMP to either intact CAP site or to two flexibly linked half-sites. In itself, this finding indicates that the protein utilizes the intrinsic mechanism. For an experimental result that further verifies this result, we used the half-site method with fluorescent-labeled CAP half-site DNA. The DNA contained the five nucleotides directly contacted by a single DNA binding domain of CAP plus additional noncontacted or weakly contacted nucleotides on either side. CAP binding to this DNA reduced its fluorescence emission by about 60%. Figure 6 shows the magnitude of the fluorescence at equilibrium as a function of the concentration of CAP in the presence and absence of 100 μM cAMP. At this cAMP concentration, the inhibitory binding sites on CAP14 should not be occupied. No detectable DNA binding was detected in the absence of cAMP, even up to 45 μM CAP. In the presence of cAMP, however, binding was readily observable at CAP concentrations above 0.1 μM. The approximately micromolar affinity for the half-site provides an upper limit15 of approximately picomolar affinity for the full-site, which is consistent with our full-site data. These results show that the intrinsic binding affinity or ability of the individual DNA binding domains of CAP changes in response to cAMP, and hence, that CAP utilizes the intrinsic mechanism.
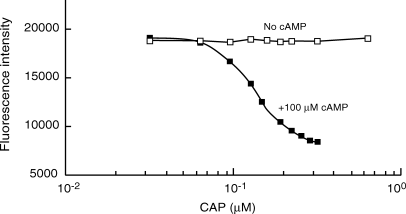
CAP binding to its DNA half-site in the presence and absence of cAMP.
Discussion
Our abilities to use computation to predict the structures of proteins, to design new proteins, and to predict the properties of proteins is developing, and promises to advance significantly in the future. One attractive area for prediction of function will be allostery. To compare prediction with experiment, it is necessary that we know more about allosteric regulation. Surprisingly, even in the case of well-studied proteins like TetR and CAP, definitive experimental evidence as to allosteric mechanism does not yet exist. The experiments described in this article were designed to alleviate this deficiency.
TetR and CAP could alter their DNA binding affinities in response to tetracycline or cAMP respectively, by the intrinsic mechanism, in which the intrinsic DNA binding affinity of the individual DBDs changes, or by the extrinsic mechanism, in which the relative spacing or orientation of the DBDs is altered. As explained in the Results section, the presence of the two mechanisms can be identified by examining binding to either a single DNA half-site, or by examining binding to two DNA half-sites connected by a flexible linker.
For the TetR protein, we used the flexible linker approach with the two TetR operator half-sites connected by a single-stranded DNA linker of one, three, or five thymines. The affinity of TetR for the latter two DNAs was nearly the same in the presence and absence of tetracycline. The simplest explanation that is consistent with the fact that tetracycline decreases the affinity of TetR for normal tetO operators and the T1-linked half-sites but leaves the affinity for T3- and T5-linked tetO half-sites largely unchanged, is that TetR utilizes the extrinsic mechanism. If the linker connecting the operator half-sites were ideal, and the repressor mechanism were entirely extrinsic, then the repressor's affinity for the operator would be unaffected by tetracycline. The slight changes in affinity for the T3- and T5- linked DNAs are likely due to nonideality in the linkers or the result of a minor contribution from an intrinsic mechanism.
Taken together, these results show that TetR employs the extrinsic mechanism and that tetracycline controls the separation or relative orientation between its two DNA binding domains. In light of the more than 10,000-fold16, 17 effect of tetracycline on binding to the normal tet operator, the near independence on tetracycline in binding to the T3-linked and T5-linked DNA suggests that any intrinsic component in the response of TetR is small. TetR structures are known for the apo18 (PDB 1A6I), effector-bound19 (PDB 2TRT), and DNA-bound13 (PDB 1QPI), forms. Utilization of the intrinsic mechanism by TetR is suggested by the presence of small structural differences, rmsd approximately 1 Å, between the DBDs of tetracycline-bound and DNA-bound TetR which may generate appreciable differences in DNA binding energies. On the other hand, utilization of an extrinsic mechanism is suggested by the fact that in the crystal structures the DBDs are approximately 3Å further apart in tetracycline-bound TetR than in the DNA-bound TetR. As explained in the introduction, neither of these facts definitively reveals the mechanism utilized by TetR. Because of the difficulty in calculating with sufficient accuracy the relevant energy differences from structures, observed shape differences could be completely without consequence, or they could dictate mechanism of action. Similar reservations can be made with respect to several solution-based studies that are consistent with a change in the relative positioning of the DNA binding domains between the DNA-bound and inducer-bound states of TetR,20-22 or tryptophan fluorescence changes indicative of a change in the structure of the DNA binding domain.23 Another experimental study24 suggests that the tetracycline binding pocket and the DNA binding domain are in communication with each other, but does not directly address the resolution of intrinsic versus extrinsic mechanisms. An extrinsic mechanism is consistent with a recent targeted molecular dynamics study of TetR's allosteric mechanism.25
For CAP, we used binding to a single half-site to determine allosteric mechanism. This binding displayed a strong cAMP dependence, indicating that the individual DNA binding domains change their affinity for DNA in response to the binding of cAMP. That is, CAP utilizes an intrinsic allosteric mechanism.
Previously, genetic and structural studies have been used to infer the effects produced by cAMP binding to CAP. Genetic studies of CAP examining cAMP independent mutants and their suppressors suggested that cAMP binding alters the positioning of the DNA binding domains of CAP with respect to the cAMP binding domains.26-28 NMR and protein foot printing experiments indicate much the same.29-31 These domain movements appear likely to change the accessibility of the DBD to the DNA, but they also change the separation between the two DNA binding domains. Thus, it is not clear which of the allosteric mechanisms they more strongly support. On the basis of a high-resolution, 2.1 Å, structure of cAMP-CAP, Passner et al.32 speculated that the main allosteric effect of cAMP binding is on subunit or domain orientation. This conclusion was based on an interpretation of chemical protection and mutational studies in the context of their high resolution structure. However, they stated that in the absence of an apo-CAP structure, only a hypothetical discussion of the mechanistically relevant conformational changes is possible. In another physical study, the kinetics of hydrogen-deuterium exchange in the presence and absence of cAMP demonstrated a cAMP-dependent alteration in the DNA binding domain that is more easily understood as a conformational change within the domain.33 Thus, as in the case for TetR, the information available before the work reported here does not strongly support one allosteric mechanism over another. The data reported here shows that CAP utilizes the intrinsic mechanism. We should note, however, that our data does not preclude a change in the relative positioning of the DBDs of CAP in response to the presence of cAMP, rather, it says that if there is such a change, its energetic consequences are small.
When it becomes possible to easily utilize high resolution structures of proteins to calculate the energetic costs of defined protein distortions, it should be much easier to predict the allosteric mechanism employed by a protein whose relevant structures are known, that is, when the protein is free in solution, when bound to effector, and when bound to DNA. In the meantime, direct experiments designed to assay the allosteric mechanism, as were used here, are the most straightforward method for mechanism determination. Thus far, the allosteric mechanisms have been directly determined for three DNA binding proteins, AraC, extrinsic,34 and in this work, TetR, extrinsic, and CAP, intrinsic.
Methods
TetR purification
TetR(BD), referred to in this article as TetR, is a fusion of the DBD (residues 1–50) of TetR, class B, and the dimerization domain (residues 51–207) of TetR, class D.18 TetR was over expressed on the pWH610 plasmid in the E. coli strain RB791, kind gifts from Wolfgan Hillen, and purified by a modification of the method of Ettner et al.35 Cells containing TetR were grown at 37°C in YT medium36 with 100 μg/mL ampicillin to an OD600 ∼0.6, induced with 0.5 mM IPTG, and grown 3 h. Cells were pelleted by centrifugation, stored overnight at 4°C, resuspended in 40 mL 20 mM Na-phosphate pH 6.8, 50 mM NaCl, 1 mM EDTA, and 1 mM DTT. Phenylmethylsulfonyl fluoride, freshly dissolved in 95% ethanol, was added to a final concentration of 0.1 mM. The resuspended cells were lysed in an Avestin Emulsiflex C3 cell homogenizer, centrifuged at 10,000g for 30 min at 4°C. The supernatant was loaded at 1 mL/min on a 5 mL Hitrap Heparin HP column, Amersham Biosciences, and eluted at 250–350 mM NaCl in a 60 mL gradient of from 150 to 450 mM NaCl in 20 mM Na-phosphate pH 6.8 buffer. The TetR-containing fractions were pooled and diluted 4–6-fold with 20 mM Na-phosphate pH 6.8, loaded at 1 mL/min on a MonoS HR 10/10 column, Pharmacia, and eluted at 200–300 mM NaCl in a 60 mL gradient from 150 to 450 mM NaCl in the Na-phosphate buffer. Eluted TetR was >95% pure as assessed by SDS-PAGE gel stained with Coomassie blue. TetR concentration was calculated using an extinction coefficient of 17,900 M−1 cm−1 per monomer.37 The activity of TetR was determined by titration against known concentrations of tetO in 10 mM Tris-acetate, pH 7.4, 50 mM NaCl, 0.5 mM magnesium acetate, 5% glycerol, 0.05% NP-40, and 0.01 mg/mL BSA, using electrophoretic mobility shift assays. The purified TetR bound specifically to tetO DNA in the absence of tetracycline, but did not bind in its presence. Purified TetR was stored at 4°C. Over 40 mg of purified TetR was obtained from the cells grown in 2 L.
Purification of CAP
The Escherichia coli CAP gene was subcloned from the plasmid pYZCRP38 into the multiple cloning region of pET3d between the NcoI and BamH1 sites. The construct was verified by DNA sequencing. After transformation into BL21(DE3) cells, CAP was purified from 1 L of cells grown in YT medium induced for 3 h with 1 mM IPTG that was added at an OD550 of 0.6 to 0.8. Cells were lysed by three passages through a French press in 20 mM Na-phosphate, pH 7, 200 mM NaCl, 2 mM EDTA, 0.5 mM 2-mercaptoethanol, 5% glycerol buffer and centrifuged at 30,000g for 30 min. After dialysis into 20 mM Tris, pH 8, the supernatant was applied to a MonoQ, Pharmacia, column equilibrated in the same buffer. CAP eluted at about 100 mM NaCl. DNA in the pooled fractions was removed by precipitation by bringing the NaCl concentration to 0.4M and adding polymine P to a concentration of 0.02%.39 The supernatant after centrifugation at 10,000g for 20 min was chromatographed on a MonoS column in 20 mM MES, pH 6 buffer. CAP eluted at about 700 mM NaCl and was greater than 98% pure.
DNA oligonucleotides
DNA oligonucleotides were from either MWG Biotech AG or Integrated DNA Technologies. When necessary, contaminants were removed by chromatography on a MonoQ column, as described previously.40 The major contact bases are underlined. Labeled full-site tetO was 5′-Cy5-GTG CTG GCC TAT CAA TGA TAG GGT GCT GG annealed to its complement. Unlabeled full-site competitor DNA was GTG CTT CCC TAT CAG TGA TAG AGT GCT GG annealed to its complement. Linked tetO half-sites were 5′-Cy5-GCT CGG CTG GAC TCT ATC A and 5′-TGA TAG GGA GTG GTC GCA annealed to CTG CGA CCA CTC CCT ATC A (Tn) TGA TAG AGT CCA GCC GAG C, where n is 1 in T1-linked, 3 in T3-linked, or 5 in T5-linked tetO half-sites. Unlabeled T5-linked DNA used as competitor was the same sequence. Non-specific DNA was made by annealing 5′-Tamara-CAG CTT AGC ATT TTT ATC CAT AGC TAC to its complement.
CAP DNA half-site used in this study was made by annealing 5′-TAMRA-ATG TCA CAT TAA TTG CGT TGC G-3′ to its complement. The sequence is that used in the determining the X-ray structure.14 The symmetry center corresponding to the full CAP palindrome is at the 5′ end of the half-site. The major contact bases are underscored.
A 210-bp full CAP site DNA was produced by PCR amplification of the CAP binding site found in the pBAD-GFP vector. The PCR primers provided for amplification of a site spanning positions 1055 through 1264 of the vector. The 40-bp CAP binding site is centered at a position 72 bp from one end. The amplified DNA was purified using a QIAGEN QIAquick PCR purification kit. Concentration was determined from the absorbance at 260 nm assuming an extinction coefficient of 50 μg/OD.
TetR binding assays
Tetracycline in 0.1M HCl, (ε355 = 13,320 M−1 cm−1)41 was adjusted to 100 mM in buffer containing 30 mM Mg-acetate. Binding buffer contained 10 mM Tris-acetate, pH 7.4, 0.5 mM magnesium acetate, 5% glycerol, 0.05% NP-40, 0.01 mg/mL BSA, and between 3.5 and 200 mM NaCl. Full-site competitor DNA and T5-linked competitor DNAs were used for experiments in the absence of tetracycline and presence of tetracyline, respectively. After addition of competitor DNA and dilution, the samples were loaded on the gel within 25 sec. Gel electrophoretic separation of protein-bound and free DNA was as described previously42 except that the electrophoresis was performed at 4°C, which maintained the temperature of the running buffer below 15°C. Fluorescent DNA bands were imaged with a Typhoon 9410 Variable Mode Imager and quantified with ImageJ.43 Details for the experimental conditions used for TetR binding to full-site and linked half-sites DNAs are described below.
Gel assay of TetR binding to full-site tetO
TetR was used at 25 nM, Cy5-labeled full-site tetO at 10 nM, and NaCl was at 150 mM in the presence or absence of 10 μM tetracycline. After 10 min incubation at 4°C, 1 μL of the incubation mix was added to 24 μL of 4.4 mM NaCl binding buffer, with or without 10 μM tetracycline, and containing 1.7 μM unlabeled competitor DNA.
Gel assay of TetR binding to linked half-sites
TetR, at concentrations ranging from 10−11 to 10−6 M, was mixed with 1.6 nM of Cy5 labeled T1-linked, T3-linked, or T5-linked DNAs in the presence and absence of 100 μM tetracycline in 200 mM NaCl binding buffer. After at least a 10 min incubation at room temperature, 2 μL of the incubation mix was added to 25 μL of 3.5 mM NaCl binding buffer, with or without 100 μM tetracycline, and containing 1.6 μM unlabeled competitor DNA. The data from three to five separate titrations were normalized, combined, and fit to the Langmuir binding isotherm.
Gel assay of CAP binding
Various concentrations of CAP were incubated with unlabeled 210 bp full-site, 71 nM, in the presence of 100 μM cAMP in buffer containing 10 mM HEPES, pH 7.4, 50 mM KCl, 0.1 mM EDTA, 2.5 mM DTE, 100 μg/mL BSA, and 5% glycerol until equilibrium had been achieved. Samples were subjected to gel electrophoresis as described above and the gel was visualized by staining with ethidium bromide and photographed.
Fluorescence assay of CAP binding
Various concentrations of CAP were incubated with TAMRA-labeled half-site, 10 nM, in the presence or absence of 100 μM cAMP in buffer containing 10 mM HEPES, pH 7.4, 50 mM KCl, 0.1 mM EDTA, 2.5 mM DTE, 100 μg/mL BSA, and 5% glycerol until equilibrium had been achieved. The 2.5 mL sample was excited at 558 nm and emission at 578 nm was integrated for 30 s.
Acknowledgements
We thank Wolfgang Hillen for the pWH610 plasmid and RB791 strain, Stephanie Dirla, and Katie Frato for assistance and comments on the article.




